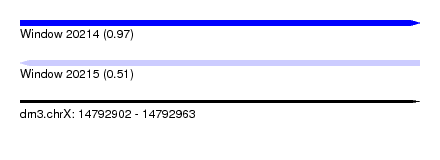
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,792,902 – 14,792,963 |
| Length | 61 |
| Max. P | 0.974811 |

| Location | 14,792,902 – 14,792,963 |
|---|---|
| Length | 61 |
| Sequences | 8 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 86.66 |
| Shannon entropy | 0.26110 |
| G+C content | 0.43410 |
| Mean single sequence MFE | -14.44 |
| Consensus MFE | -10.08 |
| Energy contribution | -10.66 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
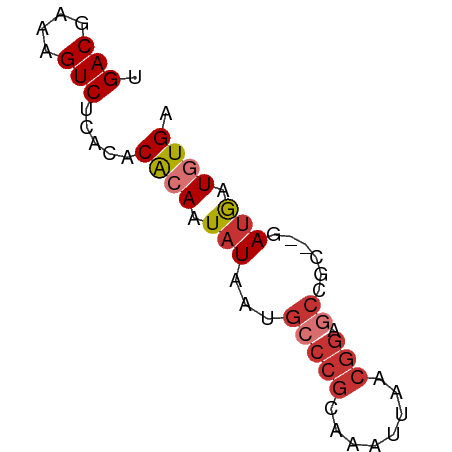
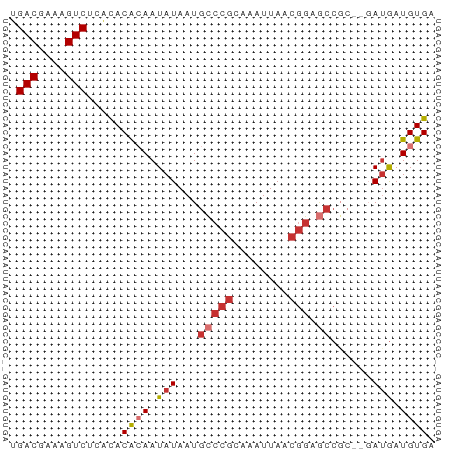
>dm3.chrX 14792902 61 + 22422827 UGACGAAAGUCUCACACACAAUAUAAUGCCCGCAAAUUAACGGAGCCGCGCGAUGAUGUGA .(((....))).....((((.(((..((((((........)))....)))..))).)))). ( -14.70, z-score = -1.63, R) >droPer1.super_63 267344 54 - 372969 UGACGAAAGUC--GUACGAAAUAUAAUGCCCGAAAAUUAUCGGAGCC-----AUGAUGUGA .(((....)))--.......((((.((((((((......)))).).)-----)).)))).. ( -13.50, z-score = -3.01, R) >dp4.chrXL_group1a 5838919 54 + 9151740 UGACGAAAGUC--GCACAAAAUAUAAUGCCCGAAAAUUAUCGGAGCC-----AUGAUGUGA .(((....)))--.((((..((.....((((((......)))).)).-----))..)))). ( -14.20, z-score = -3.31, R) >droAna3.scaffold_13335 667610 57 + 3335858 UGACGAAAGUC--CCACGCAAUAUAAUGCCCGCAAAUUAAAAAAAAAGC--GACAAUGUGA .(((....)))--.((((((......))).(((..............))--).....))). ( -11.14, z-score = -3.48, R) >droEre2.scaffold_4690 6541394 59 - 18748788 UGACGAAAGUCUCACACACAAUAUAAUGUCCGCAAAUUAACGGAUCCGC--GAUGAUGUGA .(((....))).....((((.(((...(((((........)))))....--.))).)))). ( -15.20, z-score = -2.20, R) >droYak2.chrX 9004697 59 + 21770863 UGACGAAAGUCUCACACACAAUAUAAUGCCCGCAAAUUAACGGAGCCGC--GAUGAUGUGA .(((....))).....((((.(((...(((((........))).))...--.))).)))). ( -15.60, z-score = -2.73, R) >droSec1.super_32 233318 59 + 562834 UGACGAAAGUCUCACACACAAUAUAAUGCCCGCAAAUUAACGGAGCCGC--GAUGAUGUGA .(((....))).....((((.(((...(((((........))).))...--.))).)))). ( -15.60, z-score = -2.73, R) >droSim1.chrX 11423938 59 + 17042790 UGACGAAAGUCUCACACACAAUAUAAUGCCCGCAAAUUAACGGAGCCGC--GAUGAUGUGA .(((....))).....((((.(((...(((((........))).))...--.))).)))). ( -15.60, z-score = -2.73, R) >consensus UGACGAAAGUCUCACACACAAUAUAAUGCCCGCAAAUUAACGGAGCCGC__GAUGAUGUGA .(((....)))...((((..((.....(((((........))).))......))..)))). (-10.08 = -10.66 + 0.58)

| Location | 14,792,902 – 14,792,963 |
|---|---|
| Length | 61 |
| Sequences | 8 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 86.66 |
| Shannon entropy | 0.26110 |
| G+C content | 0.43410 |
| Mean single sequence MFE | -14.62 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
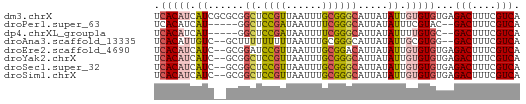
>dm3.chrX 14792902 61 - 22422827 UCACAUCAUCGCGCGGCUCCGUUAAUUUGCGGGCAUUAUAUUGUGUGUGAGACUUUCGUCA ((((((((.......((.((((......)))))).......)).))))))(((....))). ( -17.34, z-score = -1.45, R) >droPer1.super_63 267344 54 + 372969 UCACAUCAU-----GGCUCCGAUAAUUUUCGGGCAUUAUAUUUCGUAC--GACUUUCGUCA .........-----.((.((((......))))))..............--(((....))). ( -10.80, z-score = -2.09, R) >dp4.chrXL_group1a 5838919 54 - 9151740 UCACAUCAU-----GGCUCCGAUAAUUUUCGGGCAUUAUAUUUUGUGC--GACUUUCGUCA .((((..((-----(((.((((......))))))....)))..)))).--(((....))). ( -12.30, z-score = -2.09, R) >droAna3.scaffold_13335 667610 57 - 3335858 UCACAUUGUC--GCUUUUUUUUUAAUUUGCGGGCAUUAUAUUGCGUGG--GACUUUCGUCA ((((....((--((..............))))(((......)))))))--(((....))). ( -11.44, z-score = -1.81, R) >droEre2.scaffold_4690 6541394 59 + 18748788 UCACAUCAUC--GCGGAUCCGUUAAUUUGCGGACAUUAUAUUGUGUGUGAGACUUUCGUCA ((((((..((--((((((......))))))))(((......)))))))))(((....))). ( -15.60, z-score = -1.44, R) >droYak2.chrX 9004697 59 - 21770863 UCACAUCAUC--GCGGCUCCGUUAAUUUGCGGGCAUUAUAUUGUGUGUGAGACUUUCGUCA ((((((((..--...((.((((......)))))).......)).))))))(((....))). ( -16.50, z-score = -1.72, R) >droSec1.super_32 233318 59 - 562834 UCACAUCAUC--GCGGCUCCGUUAAUUUGCGGGCAUUAUAUUGUGUGUGAGACUUUCGUCA ((((((((..--...((.((((......)))))).......)).))))))(((....))). ( -16.50, z-score = -1.72, R) >droSim1.chrX 11423938 59 - 17042790 UCACAUCAUC--GCGGCUCCGUUAAUUUGCGGGCAUUAUAUUGUGUGUGAGACUUUCGUCA ((((((((..--...((.((((......)))))).......)).))))))(((....))). ( -16.50, z-score = -1.72, R) >consensus UCACAUCAUC__GCGGCUCCGUUAAUUUGCGGGCAUUAUAUUGUGUGUGAGACUUUCGUCA .(((((.((......((.((((......)))))).....)).)))))...(((....))). ( -9.90 = -10.10 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:46 2011