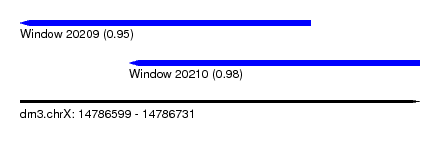
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,786,599 – 14,786,731 |
| Length | 132 |
| Max. P | 0.981255 |

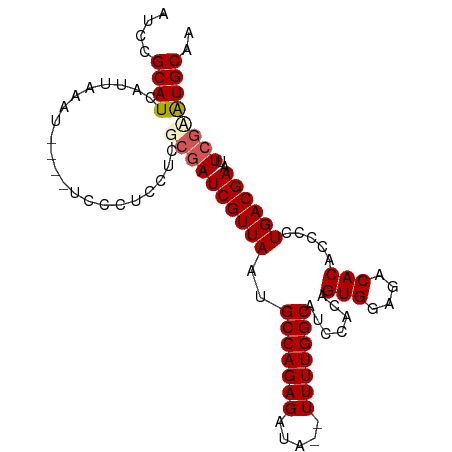
| Location | 14,786,599 – 14,786,695 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.74 |
| Shannon entropy | 0.29111 |
| G+C content | 0.44321 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14786599 96 - 22422827 UGCCAGAGAUAUUUUUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGCAUGCAAGCUGAAAAGCGGAUAUCAUUUUAUGGACCAAAAAUA (((((((((...)))))))))((((..(((....)))...((((.((....)))).....(((....)))))...........))))......... ( -25.50, z-score = -2.61, R) >droSim1.chrX_random 3882543 92 - 5698898 -GCCAGAGAUAU--UUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGUGUGCAAGCUG-CAAGCGGAUAUCAAUUUAUGGAACAUAAAUG -(((((((...)--)))))).((((..(((....))).........((((((((.(((.....)-)).)))))).))......))))......... ( -27.20, z-score = -2.64, R) >droSec1.super_32 227153 92 - 562834 -GCCAGAGAUAU--UUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGUAUGCAAGCUG-CAAGCGGAUAACAAUUUAUGGAACAUAAAUA -(((((((...)--)))))).((((..(((....)))...........((((((.(((.....)-)).)))))).........))))......... ( -26.20, z-score = -3.34, R) >droYak2.chrX 8998333 77 - 21770863 UGCCAGAGUUAU--UUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGAAUGCAAACUG-CAAGUAGUUAGCUGU---------------- ((((((((...)--)))))))......(((....)))........((....))...((.(((((-....))))).))...---------------- ( -19.40, z-score = -1.25, R) >droEre2.scaffold_4690 6535108 93 + 18748788 UGCCAGAGAUAU--UUUGGCAUUCACAGUGGAGACACACCCCUGACGAAUUCGAAUGCAAACUG-CAAGCGGAUAGCAGUUUAUGGAACAUAAAUA ((((((((...)--)))))))((((..(((....)))....(((.(..(((((..(((.....)-))..))))).))))....))))......... ( -24.70, z-score = -2.67, R) >consensus UGCCAGAGAUAU__UUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGAAUGCAAGCUG_CAAGCGGAUAGCAAUUUAUGGAACAUAAAUA .((((((.......))))))((((...(((....))).......................(((....)))))))...................... (-16.34 = -16.54 + 0.20)

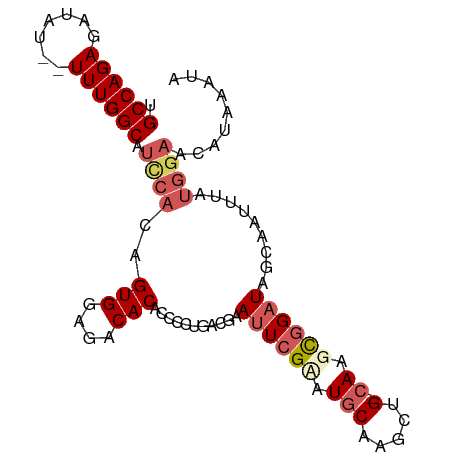
| Location | 14,786,635 – 14,786,731 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 93.56 |
| Shannon entropy | 0.11156 |
| G+C content | 0.46836 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -21.29 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14786635 96 - 22422827 AUCCGCAUCAUUAAAU----UCCCUCCUCGCGAUCGUUAAUGCCAGAGAUAUUUUUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGCAUGCAA ....((((........----.........((((((((((((((((((((...)))))))))).....(((....))).....))))))..)))))))).. ( -27.62, z-score = -3.71, R) >droSim1.chrX_random 3882578 93 - 5698898 AUCCGCAUCAUUAAAU----UCCCUCCUCGCGAUCGUUAA-GCCAGAGAUA--UUUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGUGUGCAA ....(((.........----........(((((((((((.-(((((((...--))))))).......(((....))).....))))))..)))))))).. ( -24.52, z-score = -2.42, R) >droSec1.super_32 227188 93 - 562834 AUCCGCAUCAUUAAAU----UCCAUCCUCGCGAUCGUUAA-GCCAGAGAUA--UUUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGUAUGCAA ....((((........----.........((((((((((.-(((((((...--))))))).......(((....))).....))))))..)))))))).. ( -23.72, z-score = -2.56, R) >droYak2.chrX 8998352 98 - 21770863 AUCCGCAUCAUUAAAUUCCCUCCCUCCUCGCAAUCGUUAAUGCCAGAGUUA--UUUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGAAUGCAA ....((((...................(((...(((((((((((((((...--))))))))).....(((....))).....))))))...))))))).. ( -23.40, z-score = -2.68, R) >droEre2.scaffold_4690 6535143 94 + 18748788 AUCCGCAUCAUUAAAU----UCCCUCCUCGGGAUCGUUAAUGCCAGAGAUA--UUUUGGCAUUCACAGUGGAGACACACCCCUGACGAAUUCGAAUGCAA ....(((((.......----((((.....))))(((((((((((((((...--))))))))).....(((....))).....))))))....).)))).. ( -29.60, z-score = -3.42, R) >consensus AUCCGCAUCAUUAAAU____UCCCUCCUCGCGAUCGUUAAUGCCAGAGAUA__UUUUGGCAUCCACAGUGGAGACACACCCCUGACGAAUUCGAAUGCAA ....((((.....................((((((((((..(((((((.....))))))).......(((....))).....))))))..)))))))).. (-21.29 = -21.85 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:41 2011