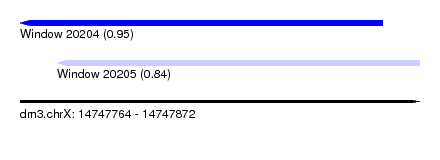
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,747,764 – 14,747,872 |
| Length | 108 |
| Max. P | 0.954622 |

| Location | 14,747,764 – 14,747,862 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 58.26 |
| Shannon entropy | 0.81828 |
| G+C content | 0.42399 |
| Mean single sequence MFE | -20.36 |
| Consensus MFE | -7.52 |
| Energy contribution | -6.34 |
| Covariance contribution | -1.17 |
| Combinations/Pair | 2.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
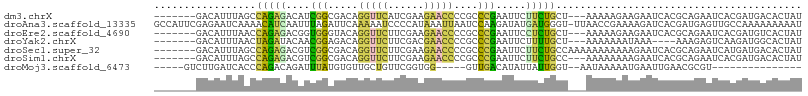
>dm3.chrX 14747764 98 - 22422827 -------GACAUUUAGCCAGAGACAUCGGCGACAGGUUCAUCGAAGAACCCCGCCCGAAUUCUUCUGCU---AAAAAGAAGAAUCACGCAGAAUCACGAUGACACUAU -------................((((((((...(((((......))))).)))).((.(((((((...---....)))))))))............))))....... ( -25.00, z-score = -2.97, R) >droAna3.scaffold_13335 619854 107 - 3335858 GCCAUUCGAGAAUCAAAACAUCAAUUUAGAUUCAAAAAUCCCCAUAAAUUAAUCCAAGAUAUGAUGGGU-UUAACCGAAAAGAUCACGAUGAGUUGCCAAAAAAAAAU ((.(((((.(((((.(((......))).))))).......(((((..(((.......)))...))))).-...................))))).))........... ( -14.10, z-score = -0.54, R) >droEre2.scaffold_4690 6497488 98 + 18748788 -------GACAUUUAACCAGAGACGGUGGGUACAGGUUCUUCGAAGAACCCCGCCCGAAUUCCUCUGCU---AAAAAGAAGAAUCACGCAGAAUCACGAUGUCACUAU -------((((((....(((((....(((((...((((((....))))))..))))).....)))))..---........((.((.....)).))..))))))..... ( -27.00, z-score = -2.43, R) >droYak2.chrX 8960372 94 - 21770863 -------GACAUUUAACUAGAUACAACGGAGACAGGUUCUUCGACGAACCCCGCCCGAAUUCUUUUGCU---AAAAAAAUAAA----AAAGAGUCAAGAUGGCACUAU -------..(((((............(((.(...(((((......)))))...)))).((((((((...---..........)----)))))))..)))))....... ( -17.22, z-score = -1.38, R) >droSec1.super_32 189163 101 - 562834 -------GACAUUUAGCCAGAGACGUCGGCGACAGGUUCUUCGAAGAACCCCGCCCGAAUUCUUCUGCCAAAAAAAAAAAGAAUCACGCAGAAUCAUGAUGACACUAU -------................((((((((...((((((....)))))).)))).......((((((...................))))))....))))....... ( -21.81, z-score = -1.55, R) >droSim1.chrX 16079112 98 - 17042790 -------GACAUUUAGCCAGAGACGUCGGCGACAGGUUCUUCGAAGAACCCCGCCCGAAUUCUUCUGCC---AAAAAAAAGAAUCACGCAGAAUCACGAUGACACUAU -------................((((((((...((((((....)))))).)))).......((((((.---...............))))))....))))....... ( -22.29, z-score = -1.65, R) >droMoj3.scaffold_6473 14953414 81 + 16943266 -----GUCUUGAUCACCCAGACAGAUUUAUGUGUUGCUGUUCGGUGG-----GUUGACAUAUUAUUGGU--AAUAAAAAUGAAUUGAACGCGU--------------- -----((((.(.....).)))).(((((((.((((((((....(((.-----.....))).....))))--))))...)))))))........--------------- ( -15.10, z-score = 0.46, R) >consensus _______GACAUUUAACCAGAGACAUCGGCGACAGGUUCUUCGAAGAACCCCGCCCGAAUUCUUCUGCU___AAAAAAAAGAAUCACGCAGAAUCACGAUGACACUAU .................(((((....(((.....(((((......)))))....))).....)))))......................................... ( -7.52 = -6.34 + -1.17)

| Location | 14,747,774 – 14,747,872 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 60.54 |
| Shannon entropy | 0.78731 |
| G+C content | 0.43037 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -6.73 |
| Energy contribution | -7.41 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
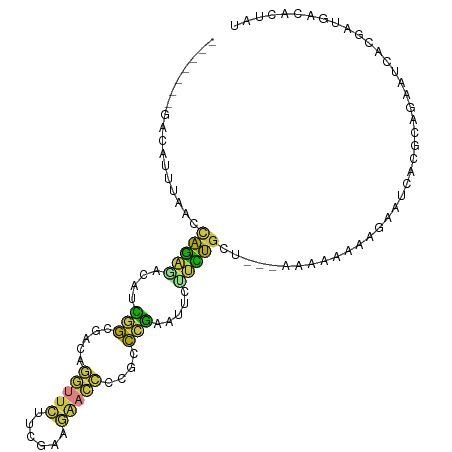
>dm3.chrX 14747774 98 - 22422827 GAUCUGGUUUGACAU--UUAGCCAGAGAC--AUCGGCGACAGGUUCAUCGAAGAACCCCGCCCGAAUUCUUCUGCUA---AAAAGAAGAAUCACGCAGAAUCACG ..(((((((......--..)))))))((.--.((((((...(((((......))))).)))).((.(((((((....---...))))))))).....)).))... ( -29.60, z-score = -3.17, R) >droAna3.scaffold_13335 619864 105 - 3335858 GAUCUGUUGCCAUUCGAGAAUCAAAACAUCAAUUUAGAUUCAAAAAUCCCCAUAAAUUAAUCCAAGAUAUGAUGGGUUUAACCGAAAAGAUCACGAUGAGUUGCC (((((.......((((.(((((.(((......))).))))).......(((((..(((.......)))...)))))......)))).)))))............. ( -15.80, z-score = 0.09, R) >droEre2.scaffold_4690 6497498 98 + 18748788 GAUUUGGUUUGACAU--UUAACCAGAGAC--GGUGGGUACAGGUUCUUCGAAGAACCCCGCCCGAAUUCCUCUGCUA---AAAAGAAGAAUCACGCAGAAUCACG (((((.((.(((...--.....(((((..--..(((((...((((((....))))))..))))).....)))))((.---...)).....))).)).)))))... ( -25.60, z-score = -1.40, R) >droYak2.chrX 8960382 94 - 21770863 GAUCUGGUUUGACAU--UUAACUAGAUAC--AACGGAGACAGGUUCUUCGACGAACCCCGCCCGAAUUCUUUUGCU----AAAAAAAUAAAAA---AGAGUCAAG .((((((((......--..))))))))..--..(((.(...(((((......)))))...)))).((((((((...----..........)))---))))).... ( -22.02, z-score = -2.37, R) >droSec1.super_32 189173 101 - 562834 GAUCUGGUUUGACAU--UUAGCCAGAGAC--GUCGGCGACAGGUUCUUCGAAGAACCCCGCCCGAAUUCUUCUGCCAAAAAAAAAAAGAAUCACGCAGAAUCAUG ..(((((((......--..)))))))((.--.((((((...((((((....)))))).)).))))..))((((((...................))))))..... ( -28.21, z-score = -2.50, R) >droSim1.chrX 16079122 98 - 17042790 GAUCUGGUUUGACAU--UUAGCCAGAGAC--GUCGGCGACAGGUUCUUCGAAGAACCCCGCCCGAAUUCUUCUGCCA---AAAAAAAGAAUCACGCAGAAUCACG ..(((((((......--..)))))))((.--.((((((...((((((....)))))).)).))))..))((((((..---..............))))))..... ( -28.39, z-score = -2.45, R) >droMoj3.scaffold_6473 14953423 80 + 16943266 --CUUGUCUUGUCUUGAUCACCCAGACAG--AUUUAUGUGUUGCUGUUCGGUGG-----GUUGACAUAUUAUUGGUAA--UAAAAAUGAAU-------------- --......((((((.(.....).))))))--((((((.((((((((....(((.-----.....))).....))))))--))...))))))-------------- ( -14.60, z-score = 0.20, R) >consensus GAUCUGGUUUGACAU__UUAGCCAGAGAC__AUCGGCGACAGGUUCUUCGAAGAACCCCGCCCGAAUUCUUCUGCUA___AAAAAAAGAAUCACGCAGAAUCACG ..(((((((..........))))))).......(((.....(((((......)))))....)))......................................... ( -6.73 = -7.41 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:37 2011