| Sequence ID | dm3.chrX |
|---|---|
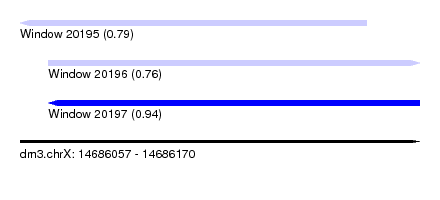
| Location | 14,686,057 – 14,686,170 |
| Length | 113 |
| Max. P | 0.938942 |

| Location | 14,686,057 – 14,686,155 |
|---|---|
| Length | 98 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.20 |
| Shannon entropy | 0.50635 |
| G+C content | 0.63123 |
| Mean single sequence MFE | -39.91 |
| Consensus MFE | -16.25 |
| Energy contribution | -17.15 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.792928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
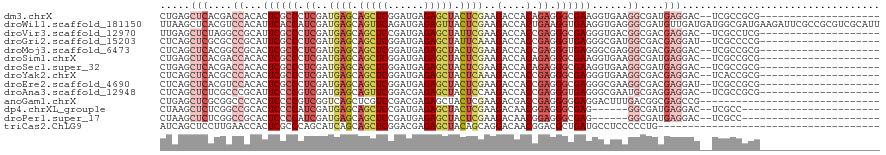
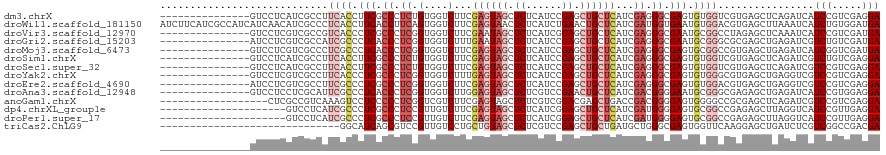
>dm3.chrX 14686057 98 - 22422827 CUGAGCUCACGACCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACAGAGGGCGAAGGUGAAGGCGAUGAGGAC--UCGCCGCG-------------------- ....((.......(((..((((((((...(((.(((((......))))).)))(.....)...))))))))..)))..(((((.......--))))))).-------------------- ( -41.20, z-score = -2.78, R) >droWil1.scaffold_181150 856708 120 + 4952429 UUAAGCUCACGUCCACAUUCACCAUCGAUGAGCAGUUCAGAUGAGAGUUACUCGAAGACCACUGAAGGUGAAGGUGAGGGCGAUGUUGAUGAUGGCGAUGAAGAUUCGCCGCGUCGCAUU ....((...(((((....(((((.((((..(((..(((....))).)))..))))..(((......)))...)))))))))).....((((.((((((.......))))))))))))... ( -37.90, z-score = -1.12, R) >droVir3.scaffold_12970 11501733 98 - 11907090 UUGAGCUCUAGGCCGCAUUCGCCCUCGAUGAGCAGCUCCGAUGAGAGCUAUUCGAAGACCACCGAGGGCGAGGGUGACGGCGACGAGGAC--UCGCCUCG-------------------- ...........((((((((((((((((..(((.(((((......))))).)))(.....)..)))))))))..))).))))..(((((..--...)))))-------------------- ( -41.50, z-score = -1.68, R) >droGri2.scaffold_15203 3484794 98 - 11997470 CUCAGCUCGCGCCCGCAUUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUAUUCAAAGACCACCGAGGGUGAGGGCGAUGGCGACGAGGAU--UCGCCCCG-------------------- .....(((((((((((.((((((((((.((((.(((((......))))).))))........)))))))))).)))..)))).))))...--........-------------------- ( -43.50, z-score = -2.31, R) >droMoj3.scaffold_6473 7190556 98 + 16943266 CUCAGCUCACGGCCGCACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACCGAGGGUGAGGGCGAGGGCGACGAGGAC--UCGCCGCG-------------------- ....((.....))(((.((((((((((..(((.(((((......))))).)))(.....)..))))))))))((((((..(.....)..)--))))))))-------------------- ( -47.20, z-score = -2.17, R) >droSim1.chrX 11336994 98 - 17042790 CUGAGCUCACGACCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACAGAGGGCGAAGGUGAAGGCGAUGAGGAC--UCGCCGCG-------------------- ....((.......(((..((((((((...(((.(((((......))))).)))(.....)...))))))))..)))..(((((.......--))))))).-------------------- ( -41.20, z-score = -2.78, R) >droSec1.super_32 133377 98 - 562834 CUGAGCUCACGACCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACAGAGGGCGAAGGUGAAGGCGAUGAGGAC--UCGCCGCG-------------------- ....((.......(((..((((((((...(((.(((((......))))).)))(.....)...))))))))..)))..(((((.......--))))))).-------------------- ( -41.20, z-score = -2.78, R) >droYak2.chrX 8905552 98 - 21770863 CUCAGCUCACGCCCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCAAAGACCACCGAGGGCGAGGGUGAAGGCGACGAGGAC--UCACCGCG-------------------- ....((...(((((((.((((((((((.((((.(((((......))))).))))........)))))))))).)))..))))....((..--...)))).-------------------- ( -44.70, z-score = -3.61, R) >droEre2.scaffold_4690 6441584 98 + 18748788 CUCAGCUCACGUCCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACCGAGGGCGAGGGCGAAGGCGACGAGGAU--UCGCCGCG-------------------- ....((...(((((....(((((((((..(((.(((((......))))).)))(.....)..))))))))))))))..(((((.......--))))))).-------------------- ( -45.40, z-score = -2.88, R) >droAna3.scaffold_12948 203722 98 + 692136 CUCAGCUCUCGCCCGCAUUCCCCGUCGAUGAGCAGUUCGGACGAGAGCUACUCCAAGACCACCGAGGGUGAGGGCGAAUGCGAGGAGGAC--UCGCCGCG-------------------- ....((((((((((.........(((...(((.(((((......))))).)))...)))......))))))))))....(((((.....)--))))....-------------------- ( -39.96, z-score = -1.11, R) >anoGam1.chrX 8917939 90 + 22145176 CUGAGCUCGCGGCCCCACUCCCCGUCGGUCAGCUCGUCCGACGAGAGCUACUCGAAGACGACCGAGGGGGAGGACUUUGACGGCGAGCCG------------------------------ ..(.((((((.((....((((((.(((((((((((.((....))))))).(.....)..)))))).))))))......).).))))))).------------------------------ ( -43.20, z-score = -1.73, R) >dp4.chrXL_group1e 7933745 89 + 12523060 CUAAGCUCUCGGCCGCACUCCCCAUCGAUGAGCAGCUCCGAUGAGAGCUACUCGAAGACAACGGAGGGCGAG------GGCGAUGAGGAC--UCGCC----------------------- ...((..((((..(((.((((((..((..(((.(((((......))))).)))(....)..))..))).)))------.))).))))..)--)....----------------------- ( -32.10, z-score = -0.47, R) >droPer1.super_17 398730 89 + 1930428 CUAAGCUCUCGGCCGCACUCCCCAUCGAUGAGCAGCUCCGAUGAGAGCUACUCGAAGACAACGGAGGGCGAG------GGCGAUGAGGAC--UCGCC----------------------- ...((..((((..(((.((((((..((..(((.(((((......))))).)))(....)..))..))).)))------.))).))))..)--)....----------------------- ( -32.10, z-score = -0.47, R) >triCas2.ChLG9 4704447 83 - 15222296 AUCAGCUCCUUGAACCACUCGCCCAGCAUCAGCAGCUCGGACGAGAGCUACAGCAGGACAACGGACGCUGAUGCCUCCCCCUG------------------------------------- ((((((((((((..((.((((((.(((.......))).)).)))).((....)).)).))).))).))))))...........------------------------------------- ( -27.60, z-score = -2.07, R) >consensus CUCAGCUCACGGCCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACCGAGGGCGAGGGCGAAGGCGACGAGGAC__UCGCCGCG____________________ .....(((....((...((((((.((..((((.(((((......))))).))))..(....).)).))))))......))....)))................................. (-16.25 = -17.15 + 0.90)
| Location | 14,686,065 – 14,686,170 |
|---|---|
| Length | 105 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.58 |
| Shannon entropy | 0.47102 |
| G+C content | 0.60585 |
| Mean single sequence MFE | -46.79 |
| Consensus MFE | -13.48 |
| Energy contribution | -14.46 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
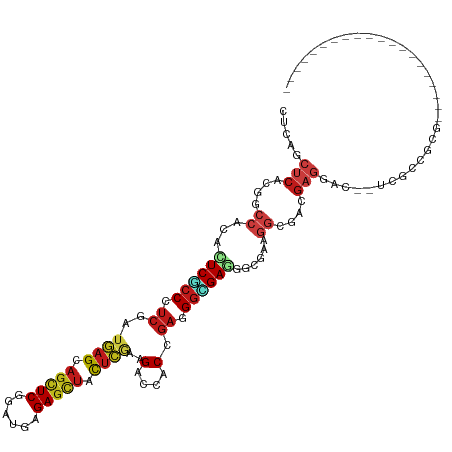
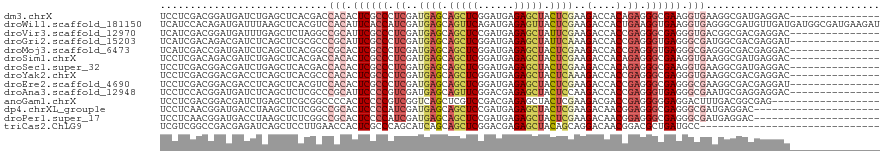
>dm3.chrX 14686065 105 + 22422827 ---------------GUCCUCAUCGCCUUCACCUUCGCCCUCUGUGGUCUUCGAGUAGCUCUCAUCCGAGCUGCUCAUCGAGGGCGAGUGUGGUCGUGAGCUCAGAUCAUCCGUCGAGGA ---------------.(((((.((((..((((.(((((((((....(....)(((((((((......)))))))))...))))))))).))))..)))).....(((.....)))))))) ( -48.40, z-score = -3.90, R) >droWil1.scaffold_181150 856723 120 - 4952429 AUCUUCAUCGCCAUCAUCAACAUCGCCCUCACCUUCACCUUCAGUGGUCUUCGAGUAACUCUCAUCUGAACUGCUCAUCGAUGGUGAAUGUGGACGUGAGCUUAAAUCAUCUGUGGAUGA ...((((.((((((..(((.(((((...........(((......)))....(((((..((......))..)))))..))))).)))..)))).))))))......(((((....))))) ( -29.10, z-score = -0.32, R) >droVir3.scaffold_12970 11501741 105 + 11907090 ---------------GUCCUCGUCGCCGUCACCCUCGCCCUCGGUGGUCUUCGAAUAGCUCUCAUCGGAGCUGCUCAUCGAGGGCGAAUGCGGCCUAGAGCUCAAAUCAUCCGUCGAUGA ---------------(..(((...((((.((...((((((((((((((........((((((....)))))))).)))))))))))).))))))...)))..)...(((((....))))) ( -44.50, z-score = -3.22, R) >droGri2.scaffold_15203 3484802 105 + 11997470 ---------------AUCCUCGUCGCCAUCGCCCUCACCCUCGGUGGUCUUUGAAUAGCUCUCAUCCGAGCUGCUCAUCGAGGGCGAAUGCGGGCGCGAGCUGAGAUCGUCUGUCGAUGA ---------------...((((.((((..(((..((.(((((((((((........(((((......))))))).))))))))).))..)))))))))))......(((((....))))) ( -44.20, z-score = -1.86, R) >droMoj3.scaffold_6473 7190564 105 - 16943266 ---------------GUCCUCGUCGCCCUCGCCCUCACCCUCGGUGGUCUUCGAGUAGCUCUCAUCCGAGCUGCUCAUCGAGGGCGAGUGCGGCCGUGAGCUGAGAUCAUCGGUCGAUGA ---------------....(((((((.((((((((((((......)))....(((((((((......)))))))))...))))))))).))((((((((.......))).)))))))))) ( -54.90, z-score = -4.18, R) >droSim1.chrX 11337002 105 + 17042790 ---------------GUCCUCAUCGCCUUCACCUUCGCCCUCUGUGGUCUUCGAGUAGCUCUCAUCCGAGCUGCUCAUCGAGGGCGAGUGUGGUCGUGAGCUCAGAUCGUCUGUCGAGGA ---------------.(((((.((((..((((.(((((((((....(....)(((((((((......)))))))))...))))))))).))))..))))...(((.....)))..))))) ( -49.60, z-score = -4.00, R) >droSec1.super_32 133385 105 + 562834 ---------------GUCCUCAUCGCCUUCACCUUCGCCCUCUGUGGUCUUCGAGUAGCUCUCAUCCGAGCUGCUCAUCGAGGGCGAGUGUGGUCGUGAGCUCAGAUCGUCCGUCGAGGA ---------------.(((((.((((..((((.(((((((((....(....)(((((((((......)))))))))...))))))))).))))..)))).....(((.....)))))))) ( -48.40, z-score = -3.69, R) >droYak2.chrX 8905560 105 + 21770863 ---------------GUCCUCGUCGCCUUCACCCUCGCCCUCGGUGGUCUUUGAGUAGCUCUCAUCCGAGCUGCUCAUCGAGGGCGAGUGUGGGCGUGAGCUGAGGUCGUCCGUCGAGGA ---------------.((((((.(((((((((.(((((((((((.......((((((((((......))))))))))))))))))))).)))(((....))))))).)).....)))))) ( -60.31, z-score = -5.42, R) >droEre2.scaffold_4690 6441592 105 - 18748788 ---------------AUCCUCGUCGCCUUCGCCCUCGCCCUCGGUGGUCUUCGAGUAGCUCUCAUCCGAGCUGCUCAUCGAGGGCGAGUGUGGACGUGAGCUGAGGUCGUCCGUCGAGGA ---------------.((((((((((.(((((.((((((((((((.(....)(((((((((......))))))))))))))))))))).))))).)))).....((....))..)))))) ( -57.70, z-score = -4.49, R) >droAna3.scaffold_12948 203730 105 - 692136 ---------------GUCCUCCUCGCAUUCGCCCUCACCCUCGGUGGUCUUGGAGUAGCUCUCGUCCGAACUGCUCAUCGACGGGGAAUGCGGGCGAGAGCUGAGAUCAUCCGUGGAGGA ---------------.((((((.(((((((.(((.((((...))))(((...((((((.((......)).))))))...)))))))))))))(((....)))............)))))) ( -47.30, z-score = -1.82, R) >anoGam1.chrX 8917942 102 - 22145176 ------------------CUCGCCGUCAAAGUCCUCCCCCUCGGUCGUCUUCGAGUAGCUCUCGUCGGACGAGCUGACCGACGGGGAGUGGGGCCGCGAGCUCAGAUCGUCCGUCGAGGA ------------------(((((.(((......((((((.((((((((((.((((.....))))..)))).....)))))).))))))...))).)))))(((.(((.....)))))).. ( -43.40, z-score = 0.06, R) >dp4.chrXL_group1e 7933750 99 - 12523060 ---------------------GUCCUCAUCGCCCUCGCCCUCCGUUGUCUUCGAGUAGCUCUCAUCGGAGCUGCUCAUCGAUGGGGAGUGCGGCCGAGAGCUUAGGUCAUCCGUUGAGGA ---------------------.(((((((((.((.(((.((((((((.....((((((((((....))))))))))..)))))))).))).)).))).......((....))..)))))) ( -44.70, z-score = -2.58, R) >droPer1.super_17 398735 99 - 1930428 ---------------------GUCCUCAUCGCCCUCGCCCUCCGUUGUCUUCGAGUAGCUCUCAUCGGAGCUGCUCAUCGAUGGGGAGUGCGGCCGAGAGCUUAGGUCAUCCGUUGAGGA ---------------------.(((((((((.((.(((.((((((((.....((((((((((....))))))))))..)))))))).))).)).))).......((....))..)))))) ( -44.70, z-score = -2.58, R) >triCas2.ChLG9 4704455 90 + 15222296 ------------------------------GGCAUCAGCGUCCGUUGUCCUGCUGUAGCUCUCGUCCGAGCUGCUGAUGCUGGGCGAGUGGUUCAAGGAGCUGAUCUCGUCGGCCGACGA ------------------------------(((..(((((((.((......)).(((((((......))))))).)))))))((((((.(((((...)))))...)))))).)))..... ( -37.80, z-score = -0.98, R) >consensus _______________GUCCUCGUCGCCUUCGCCCUCGCCCUCGGUGGUCUUCGAGUAGCUCUCAUCCGAGCUGCUCAUCGAGGGCGAGUGCGGCCGUGAGCUGAGAUCAUCCGUCGAGGA ......................(((((.((........((.....)).....((((((.((......)).))))))...)).)))))......................(((.....))) (-13.48 = -14.46 + 0.98)

| Location | 14,686,065 – 14,686,170 |
|---|---|
| Length | 105 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.58 |
| Shannon entropy | 0.47102 |
| G+C content | 0.60585 |
| Mean single sequence MFE | -42.36 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.45 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14686065 105 - 22422827 UCCUCGACGGAUGAUCUGAGCUCACGACCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACAGAGGGCGAAGGUGAAGGCGAUGAGGAC--------------- ((((((.(((.....))).(((......(((..((((((((...(((.(((((......))))).)))(.....)...))))))))..)))..)))..)))))).--------------- ( -41.40, z-score = -2.59, R) >droWil1.scaffold_181150 856723 120 + 4952429 UCAUCCACAGAUGAUUUAAGCUCACGUCCACAUUCACCAUCGAUGAGCAGUUCAGAUGAGAGUUACUCGAAGACCACUGAAGGUGAAGGUGAGGGCGAUGUUGAUGAUGGCGAUGAAGAU (((((.(((..(((.......)))(((((....(((((.((((..(((..(((....))).)))..))))..(((......)))...)))))))))).))).)))))............. ( -28.50, z-score = 0.72, R) >droVir3.scaffold_12970 11501741 105 - 11907090 UCAUCGACGGAUGAUUUGAGCUCUAGGCCGCAUUCGCCCUCGAUGAGCAGCUCCGAUGAGAGCUAUUCGAAGACCACCGAGGGCGAGGGUGACGGCGACGAGGAC--------------- (((((....)))))......(((...((((((((((((((((..(((.(((((......))))).)))(.....)..)))))))))..))).))))...)))...--------------- ( -41.50, z-score = -1.96, R) >droGri2.scaffold_15203 3484802 105 - 11997470 UCAUCGACAGACGAUCUCAGCUCGCGCCCGCAUUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUAUUCAAAGACCACCGAGGGUGAGGGCGAUGGCGACGAGGAU--------------- ..((((.....)))).....(((((((((((.((((((((((.((((.(((((......))))).))))........)))))))))).)))..)))).))))...--------------- ( -44.80, z-score = -2.54, R) >droMoj3.scaffold_6473 7190564 105 + 16943266 UCAUCGACCGAUGAUCUCAGCUCACGGCCGCACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACCGAGGGUGAGGGCGAGGGCGACGAGGAC--------------- (((((....)))))((((.((.....))(((.(((((((((..((((.(((((......))))).))))...(((......)))))))))))).)))..))))..--------------- ( -48.70, z-score = -2.98, R) >droSim1.chrX 11337002 105 - 17042790 UCCUCGACAGACGAUCUGAGCUCACGACCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACAGAGGGCGAAGGUGAAGGCGAUGAGGAC--------------- ((((((.(((.....))).(((......(((..((((((((...(((.(((((......))))).)))(.....)...))))))))..)))..)))..)))))).--------------- ( -42.10, z-score = -2.97, R) >droSec1.super_32 133385 105 - 562834 UCCUCGACGGACGAUCUGAGCUCACGACCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACAGAGGGCGAAGGUGAAGGCGAUGAGGAC--------------- ((((((.(((.....))).(((......(((..((((((((...(((.(((((......))))).)))(.....)...))))))))..)))..)))..)))))).--------------- ( -41.40, z-score = -2.52, R) >droYak2.chrX 8905560 105 - 21770863 UCCUCGACGGACGACCUCAGCUCACGCCCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCAAAGACCACCGAGGGCGAGGGUGAAGGCGACGAGGAC--------------- ((((((..((....))........(((((((.((((((((((.((((.(((((......))))).))))........)))))))))).)))..)))).)))))).--------------- ( -51.70, z-score = -4.76, R) >droEre2.scaffold_4690 6441592 105 + 18748788 UCCUCGACGGACGACCUCAGCUCACGUCCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACCGAGGGCGAGGGCGAAGGCGACGAGGAU--------------- ((((((..(((((...........)))))...((((((((((..(((.(((((......))))).)))(.....)..)))))))))).((....))..)))))).--------------- ( -51.90, z-score = -4.24, R) >droAna3.scaffold_12948 203730 105 + 692136 UCCUCCACGGAUGAUCUCAGCUCUCGCCCGCAUUCCCCGUCGAUGAGCAGUUCGGACGAGAGCUACUCCAAGACCACCGAGGGUGAGGGCGAAUGCGAGGAGGAC--------------- ((((((..((.(((.........)))))(((((((((((((...(((.(((((......))))).)))...)))((((...)))).))).))))))).)))))).--------------- ( -44.60, z-score = -2.11, R) >anoGam1.chrX 8917942 102 + 22145176 UCCUCGACGGACGAUCUGAGCUCGCGGCCCCACUCCCCGUCGGUCAGCUCGUCCGACGAGAGCUACUCGAAGACGACCGAGGGGGAGGACUUUGACGGCGAG------------------ ..((((.(((((((.((((.((.((((.........)))).)))))).))))))).)))).(((..((((((.(..((....))..)..)))))).)))...------------------ ( -41.20, z-score = 0.21, R) >dp4.chrXL_group1e 7933750 99 + 12523060 UCCUCAACGGAUGACCUAAGCUCUCGGCCGCACUCCCCAUCGAUGAGCAGCUCCGAUGAGAGCUACUCGAAGACAACGGAGGGCGAGGGCGAUGAGGAC--------------------- ((((((.(((....))...(((((((.((...((((...((..((((.(((((......))))).))))..))....)))))))))))))).)))))).--------------------- ( -40.60, z-score = -2.58, R) >droPer1.super_17 398735 99 + 1930428 UCCUCAACGGAUGACCUAAGCUCUCGGCCGCACUCCCCAUCGAUGAGCAGCUCCGAUGAGAGCUACUCGAAGACAACGGAGGGCGAGGGCGAUGAGGAC--------------------- ((((((.(((....))...(((((((.((...((((...((..((((.(((((......))))).))))..))....)))))))))))))).)))))).--------------------- ( -40.60, z-score = -2.58, R) >triCas2.ChLG9 4704455 90 - 15222296 UCGUCGGCCGACGAGAUCAGCUCCUUGAACCACUCGCCCAGCAUCAGCAGCUCGGACGAGAGCUACAGCAGGACAACGGACGCUGAUGCC------------------------------ (((((....))))).((((((((((((..((.((((((.(((.......))).)).)))).((....)).)).))).))).))))))...------------------------------ ( -34.10, z-score = -2.19, R) >consensus UCCUCGACGGACGAUCUCAGCUCACGGCCACACUCGCCCUCGAUGAGCAGCUCGGAUGAGAGCUACUCGAAGACCACCGAGGGCGAGGGCGAAGGCGACGAGGAC_______________ ............................(((.((((((.((..((((.(((((......))))).))))..(....).)).)))))).)))............................. (-19.00 = -19.45 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:31 2011