
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,657,440 – 14,657,581 |
| Length | 141 |
| Max. P | 0.798520 |

| Location | 14,657,440 – 14,657,549 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 53.81 |
| Shannon entropy | 0.73251 |
| G+C content | 0.34049 |
| Mean single sequence MFE | -17.51 |
| Consensus MFE | -7.74 |
| Energy contribution | -7.67 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
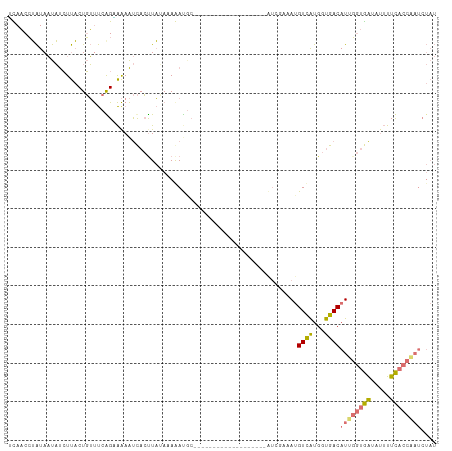
>dm3.chrX 14657440 109 + 22422827 AUAACCUAUAAUAUCUUACUGUUUCGGUAAAAUCACCUAGAAAAAUGCCAAAUAUCACAUAU--GUUAUCGAAAUGUCAUGGUGACAUAGGUGAUAUUUUCACCUAUCUAU ...............((((((...)))))).......((((....((..(((((((((.(((--(((((((........)))))))))).))))))))).))....)))). ( -22.40, z-score = -2.28, R) >droAna3.scaffold_12948 76439 84 - 692136 GUACCGGGUAUCUCAUCCCUUGCCCAGCGGAGCUAAUUUAAUAAAUGC-------------------GUGCAUACGUUGCGACAACAUCUAAAGGUCAUUUAC-------- (.((((((((..........)))))...(((((.............))-------------------(((((.....))).))....)))...))))......-------- ( -14.82, z-score = 0.64, R) >droSec1.super_32 109743 84 + 562834 UCAACCUAUAAUAUCUUACUGUUUCGGAAAAAUCACUUAUAAAGAC---------------------------AUGUCAUGGUGACAUUGGUGAUAUUUUCACCAAUCUAU ....((...((((......))))..))...............(((.---------------------------.((((.....))))(((((((.....)))))))))).. ( -13.70, z-score = -0.68, R) >droSim1.chrX 11315762 111 + 17042790 UCAACCUAUAAUAUCUUACUGUUUCAGGAAAAUCACUUAUAAAGAUGCUUAAUAUCACACAUUUGUAAUCGAAAUGUCAUGGUGACAUUGGUGAAAUUUUCACCAAUCUAU ....(((..((((......))))..)))....(((((......((((.....))))..((((((.......))))))...))))).((((((((.....)))))))).... ( -19.10, z-score = -1.13, R) >consensus UCAACCUAUAAUAUCUUACUGUUUCAGAAAAAUCACUUAUAAAAAUGC___________________AUCGAAAUGUCAUGGUGACAUUGGUGAUAUUUUCACCAAUCUAU ...........................................................................((((...))))((((((((.....)))))))).... ( -7.74 = -7.67 + -0.06)


| Location | 14,657,440 – 14,657,549 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 53.81 |
| Shannon entropy | 0.73251 |
| G+C content | 0.34049 |
| Mean single sequence MFE | -20.38 |
| Consensus MFE | -8.05 |
| Energy contribution | -7.30 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
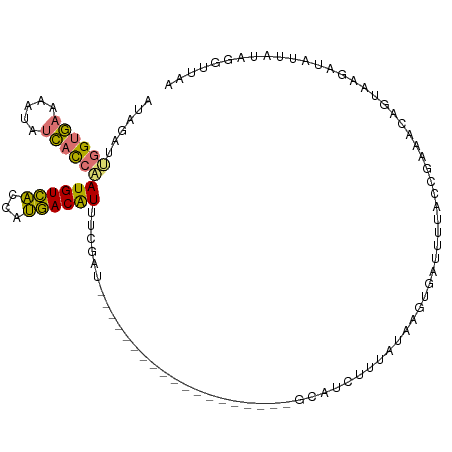
>dm3.chrX 14657440 109 - 22422827 AUAGAUAGGUGAAAAUAUCACCUAUGUCACCAUGACAUUUCGAUAAC--AUAUGUGAUAUUUGGCAUUUUUCUAGGUGAUUUUACCGAAACAGUAAGAUAUUAUAGGUUAU ....((((((((.....))))))))(((.....)))......(((((--...((((((((((.((...((((..((((....))))))))..)).)))))))))).))))) ( -27.00, z-score = -2.70, R) >droAna3.scaffold_12948 76439 84 + 692136 --------GUAAAUGACCUUUAGAUGUUGUCGCAACGUAUGCAC-------------------GCAUUUAUUAAAUUAGCUCCGCUGGGCAAGGGAUGAGAUACCCGGUAC --------.......(((..(((((((.((.(((.....)))))-------------------)))))))........((((....))))..(((........)))))).. ( -18.20, z-score = 0.22, R) >droSec1.super_32 109743 84 - 562834 AUAGAUUGGUGAAAAUAUCACCAAUGUCACCAUGACAU---------------------------GUCUUUAUAAGUGAUUUUUCCGAAACAGUAAGAUAUUAUAGGUUGA ....((((((((.....)))))))).(((((((((..(---------------------------(((((.....(.((....)))........)))))))))).)).))) ( -15.22, z-score = -0.53, R) >droSim1.chrX 11315762 111 - 17042790 AUAGAUUGGUGAAAAUUUCACCAAUGUCACCAUGACAUUUCGAUUACAAAUGUGUGAUAUUAAGCAUCUUUAUAAGUGAUUUUCCUGAAACAGUAAGAUAUUAUAGGUUGA ..((((.(((((.....)))))((((((((....((((((.......))))))))))))))....))))((((((...(((((.(((...))).))))).))))))..... ( -21.10, z-score = -0.60, R) >consensus AUAGAUUGGUGAAAAUAUCACCAAUGUCACCAUGACAUUUCGAU___________________GCAUCUUUAUAAGUGAUUUUACCGAAACAGUAAGAUAUUAUAGGUUAA ......((((((.....))))))((((((...))))))......................................................................... ( -8.05 = -7.30 + -0.75)


| Location | 14,657,473 – 14,657,581 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 60.78 |
| Shannon entropy | 0.69526 |
| G+C content | 0.34776 |
| Mean single sequence MFE | -20.94 |
| Consensus MFE | -8.39 |
| Energy contribution | -9.23 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
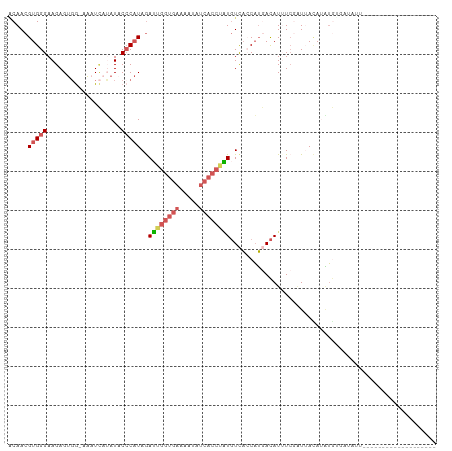
>dm3.chrX 14657473 108 - 22422827 ACAACGUGGGAAGAGUGGAAAAUCAUAUACCCAUAGAUAGGUGAAAAUAUCACCUAUGUCACCAUGACAUUUCGAUAACAUAUGUGAUAUUUGGCAUUUUUCUAGGUG-- ...((.((((((((((.(((.((((((((.......((((((((.....))))))))(((.....)))............)))))))).))).)).)))))))).)).-- ( -32.50, z-score = -3.39, R) >droEre2.scaffold_4690 6420100 90 + 18748788 ACAACGUGGGAAGAGCUA-AGAUUAAAUACCCCUAAAGU------AAUAUACCCUUUGUCAGCAUAACAUUUCUAUUA-AUAUAAUAUAAGCAUCUUU------------ .........((((((((.-..((((..(((.......))------)..........((....))..............-...))))...))).)))))------------ ( -5.60, z-score = 1.96, R) >droYak2.chrX 8883856 91 - 21770863 UCAACGUGGGAAGAGUGGAAAUUACAAUCCACAUAGAUAGGUGAAAAUAUCACCUGUUUUUACUGCAUAUGUAGGUAAUAUUAAGUAUCUU------------------- ...(((((...((.(((((........)))))..((((((((((.....))))))))))...))...)))))(((((........))))).------------------- ( -22.70, z-score = -2.45, R) >droSec1.super_32 109776 82 - 562834 GCAACGUGGGAAGAGUGG-AAAUCAUAUACCCAUAGAUUGGUGAAAAUAUCACCAAUGUCACCAUGACAUGUC-UUUAUAAGUG-------------------------- ....(((((((...((((-...........))))..((((((((.....)))))))).)).))))).......-..........-------------------------- ( -19.20, z-score = -1.11, R) >droSim1.chrX 11315795 109 - 17042790 GCAACGUGGGAAGAGUGG-AAAUCAUAUACCCAUAGAUUGGUGAAAAUUUCACCAAUGUCACCAUGACAUUUCGAUUACAAAUGUGUGAUAUUAAGCAUCUUUAUAAGUG ...((....(((((((..-((((((((((.......((((((((.....))))))))(((.....)))..............)))))))).))..)).)))))....)). ( -24.70, z-score = -1.14, R) >consensus ACAACGUGGGAAGAGUGG_AAAUCAUAUACCCAUAGAUUGGUGAAAAUAUCACCUAUGUCACCAUGACAUUUCGAUUACAUAUGUGAUAUU___________________ .....(((((...................)))))..((((((((.....))))))))..................................................... ( -8.39 = -9.23 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:27 2011