
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,656,142 – 14,656,197 |
| Length | 55 |
| Max. P | 0.655337 |

| Location | 14,656,142 – 14,656,197 |
|---|---|
| Length | 55 |
| Sequences | 6 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 68.09 |
| Shannon entropy | 0.58037 |
| G+C content | 0.34617 |
| Mean single sequence MFE | -7.50 |
| Consensus MFE | -5.17 |
| Energy contribution | -3.53 |
| Covariance contribution | -1.64 |
| Combinations/Pair | 1.86 |
| Mean z-score | -0.36 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.655337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
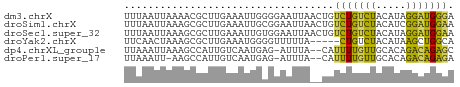
>dm3.chrX 14656142 55 - 22422827 UUUAAUUAAAACGCUUGAAAUUGGGGAAUUAACUGUCUGUCUACAUAGGAUGGGA .((((((....(.(........).).))))))...(((((((.....))))))). ( -6.20, z-score = 0.16, R) >droSim1.chrX 11314452 55 - 17042790 UUUAAUUAAAGCGCUUGAAAUUGCGGAAUUAACUGUCUGUCUACAUCGGAUGGAA .((((((....(((........))).))))))(((((((.......))))))).. ( -10.90, z-score = -1.48, R) >droSec1.super_32 108443 55 - 562834 UUUAAUUAAAGCGCUUGAAAUUGUGGAAUUAACUGUCUGUCUACAUAGGAUGGAA .((((((....(((........))).))))))...(((((((.....))))))). ( -8.20, z-score = -0.65, R) >droYak2.chrX 8882584 50 - 21770863 UUCAACUAAAGCGCUUGAAAUGGGGUUUUUA-----CUGUCUACAUAAGCUGGCA ..........(((((((...((((((....)-----))..)))..)))))..)). ( -5.50, z-score = 1.31, R) >dp4.chrXL_group1e 7905596 52 + 12523060 UUAAAUUAAAGCCAUUGUCAAUGAG-AUUUA--CAUUUUGUUGCACAGACAGAGC ..............(((((((..((-((...--.))))..))).))))....... ( -7.10, z-score = -0.63, R) >droPer1.super_17 370498 51 + 1930428 UUAAAUU-AAGCCAUUGUCAAUGAG-AUUUA--CAUUUUGUUGCACAGACAGAGA .......-......(((((((..((-((...--.))))..))).))))....... ( -7.10, z-score = -0.88, R) >consensus UUUAAUUAAAGCGCUUGAAAUUGAGGAAUUA__UGUCUGUCUACAUAGGAUGGGA ...................................(((((((.....))))))). ( -5.17 = -3.53 + -1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:25 2011