| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,633,544 – 14,633,606 |
| Length | 62 |
| Max. P | 0.869442 |

| Location | 14,633,544 – 14,633,606 |
|---|---|
| Length | 62 |
| Sequences | 9 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 75.59 |
| Shannon entropy | 0.45855 |
| G+C content | 0.36938 |
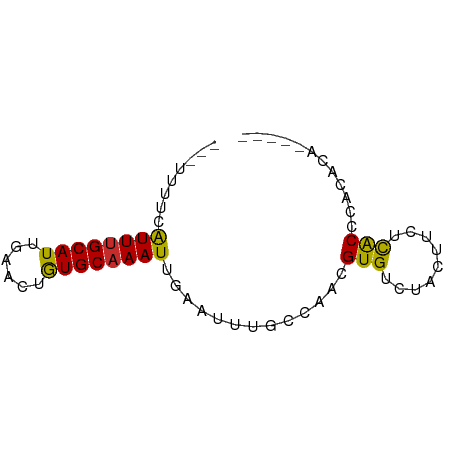
| Mean single sequence MFE | -12.09 |
| Consensus MFE | -7.22 |
| Energy contribution | -6.69 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869442 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
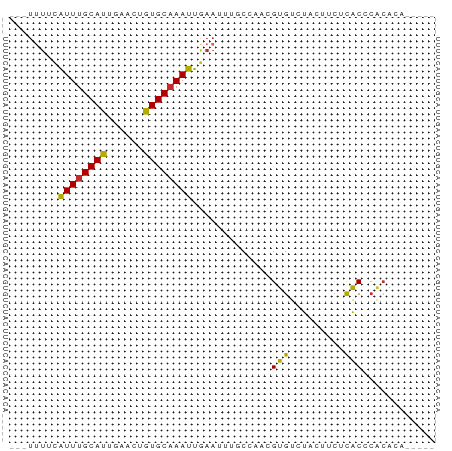
>dm3.chrX 14633544 62 - 22422827 ---UUUUCAUUUGCAUUGAACUGUGCAAAUUGAAUUUUCCAACGUGUCUACUUCUCACCCACACA----- ---..(((((((((((......))))))).)))).........((((.............)))).----- ( -11.02, z-score = -2.46, R) >droEre2.scaffold_4690 6394651 61 + 18748788 ---UUUUCAUUUGCAUUGAACUGUGC-AAUUGAAUUUUCCAACGUGUCUACUCCUCACCCGCACA----- ---..((((.((((((......))))-)).)))).........((((.............)))).----- ( -11.92, z-score = -2.81, R) >droYak2.chrX 8860052 61 - 21770863 ---UUUUCAUUUGCAUUGAACUGUGCAAAUUGAAUUUGCCAACGUGUCUAUUUCUCACCCACAU------ ---..(((((((((((......))))))).)))).........(((.............)))..------ ( -9.72, z-score = -1.26, R) >droSec1.super_32 86390 62 - 562834 ---UUUUCAUUUGCAUUGAACUGUGCAAAUUGAAUUUUCCAACGUGUCUACUUUUCACCCACACA----- ---..(((((((((((......))))))).)))).........((((.............)))).----- ( -11.02, z-score = -2.44, R) >droSim1.chrX 11292200 62 - 17042790 ---UUUUCAUUUGCAUUGAACUGUGCAAAUUGAAUUUUCCAACGUGUCUACUUCUCACCCACACA----- ---..(((((((((((......))))))).)))).........((((.............)))).----- ( -11.02, z-score = -2.46, R) >droWil1.scaffold_181150 789123 65 + 4952429 ---UUUUCAUUUGCAUUGAGCUGUGCAAAUUAAA-UUGCAAGUGUGUGUGC-ACUCACUCGCAGUCAUAA ---.....((((((((......))))))))...(-((((.((((.((....-)).)))).)))))..... ( -18.00, z-score = -1.34, R) >droVir3.scaffold_12970 460409 61 + 11907090 ---UUUCUGUUUGCAUUGAGCUGUGCAAAUUAAA-UUGCAAAUGUGUCUUGCUUAUACUAACACA----- ---....(((..(((....((..(((((......-)))))...))....)))..)))........----- ( -10.50, z-score = -0.03, R) >droMoj3.scaffold_6473 9836209 62 - 16943266 UCUGCACUGUUUGCAUUGAGCUAUGCAAAUUAAA-UUGCAAAUGUGUCUUACUUAUGCACACA------- ..((((..((((((((......))))))))....-.))))..((((((........).)))))------- ( -14.30, z-score = -0.95, R) >droGri2.scaffold_15203 8565100 59 + 11997470 ---UCUCUGUUUGCAUUGAGCUGUGCAAAUUAAA-UUGCAAAUGUGUCUUGCUCGUACACUGA------- ---((..(((..(((....((..(((((......-)))))...))....)))....)))..))------- ( -11.30, z-score = -0.42, R) >consensus ___UUUUCAUUUGCAUUGAACUGUGCAAAUUGAAUUUGCCAACGUGUCUACUUCUCACCCACACA_____ ........((((((((......)))))))).............(((.........)))............ ( -7.22 = -6.69 + -0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:20 2011