| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,631,119 – 14,631,211 |
| Length | 92 |
| Max. P | 0.996332 |

| Location | 14,631,119 – 14,631,211 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Shannon entropy | 0.43698 |
| G+C content | 0.48016 |
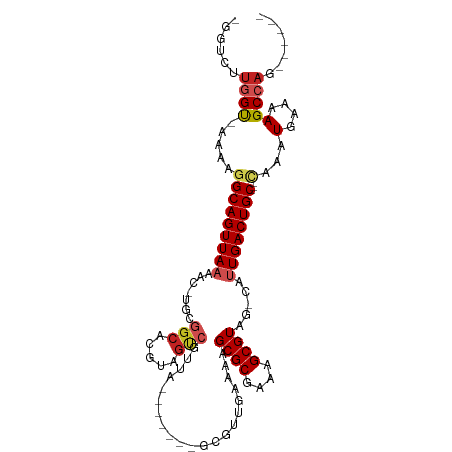
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14631119 92 - 22422827 -AAUCUUGGU-AAAAGGCAGUUAAAAC-UGUGGCACGUAGUCGUUA-------GCGUUGAAAAGCGCGAAAGCGUAG-CAUUGACUGC---CAAAUGAAAAGCCAG------ -.....((((-....(((((((((...-(((...((((..((....-------(((((....)))))))..)))).)-))))))))))---)...(....))))).------ ( -28.40, z-score = -2.32, R) >droSim1.chrX_random 3869483 92 - 5698898 -GGUCUUGGU-AAAAGGCAGUUAAAAC-UGUGGCACGUAGUCGUUA-------GCGUUGAAAAGCGCGAAAGCGUAG-CAUUGACUGC---CAAAUGAAAAGCCAG------ -((.(((..(-(...(((((((((...-(((...((((..((....-------(((((....)))))))..)))).)-))))))))))---)...))..)))))..------ ( -29.70, z-score = -2.42, R) >droSec1.super_32 84126 92 - 562834 -GGUCUUGGU-AAAAGGCAGUUAAAAC-UGUGGCACGUAGUCGUUA-------GCGUUGAAAAGCGCGAAAGCGUAG-CAUUGACUGC---CAAAUGAAAAGCCAG------ -((.(((..(-(...(((((((((...-(((...((((..((....-------(((((....)))))))..)))).)-))))))))))---)...))..)))))..------ ( -29.70, z-score = -2.42, R) >droYak2.chrX 8857732 93 - 21770863 GUUCCUUGGU-AAAAGGCAGUUAAAAC-UGUGGCACGUAGUCGUUA-------GCGUUGAAAAGCGCGAAAGCGUAG-CAUUGACUGC---CAAAUGAAAAGCCAG------ ......((((-....(((((((((...-(((...((((..((....-------(((((....)))))))..)))).)-))))))))))---)...(....))))).------ ( -28.40, z-score = -2.00, R) >droEre2.scaffold_4690 6392434 92 + 18748788 -GGUCUUGGU-AAAAGGCAGUUAAAAC-UGUGGCACGUAGUCGUUA-------GCGUUGAAAAGCGCGAAAGCGUAG-CAUUGACUGC---CAAAUGAAAAGCCAG------ -((.(((..(-(...(((((((((...-(((...((((..((....-------(((((....)))))))..)))).)-))))))))))---)...))..)))))..------ ( -29.70, z-score = -2.42, R) >droAna3.scaffold_12948 39216 91 + 692136 -GCUGUAGGU-AAAAGGCAGUUAAAAC-UGCAGCACGUAGUCGUUA-------GCGUUGAAAAGCGCGAAAGCGUAG-CAUUGACUGC---CAAAUG-AAAGCCAG------ -......(((-....(((((((((..(-(((.(((((....)))..-------(((((....)))))....))))))-..))))))))---).....-...)))..------ ( -30.10, z-score = -2.39, R) >dp4.chrXL_group1e 7875922 107 + 12523060 -CAGAGUGGU-AAAAGGCAGUUAAAAAGUGCGGCACGUAGUCGCCGCAGUUGCAUGUUGAAAAGCGCGAAAGCGUAGCCGUUGACUGC---CAAAUGAAAAGCCAGUCCAAA -..((.((((-....(((((((((....((((((........))))))(((((..(((....)))((....)))))))..))))))))---)...(....))))).)).... ( -42.70, z-score = -5.26, R) >droPer1.super_17 340932 107 + 1930428 -CAGAGUGGU-AAAAGGCAGUUAAAAACUGCGGCACGUAGUCGCCGCAGUUGCAUGUUGAAAAGCGCGAAAGCGUAGCCGUUGACUGC---CAAAUGAAAAGCCAGUCCAAA -..((.((((-....(((((((((.(((((((((........)))))))))((..(((....)))((....))...))..))))))))---)...(....))))).)).... ( -44.40, z-score = -5.35, R) >droWil1.scaffold_181150 784931 77 + 4952429 ---AGUUGGUGAAAAGGCAGUUAAAAA-CG------------GUUG-------GGA---AAACGCGCGAAAGCGUAGCCAUUGACUGC---AAAAUGAAAAGCCAG------ ---...((((......((((((((...-.(------------((((-------...---...)((((....)))))))).))))))))---..........)))).------ ( -23.19, z-score = -2.73, R) >droVir3.scaffold_12970 457280 91 + 11907090 --GCCGUAGCUAAAAGGCAGUUAAAAACUGCGGCACGUAGCCAAUG-------GUAAU-GGGCGCGCGAAAGCGUUG-CGAUGACUGC----GAAUGAAAAGCCAG------ --(((((.........(((((.....)))))(((.....))).)))-------))...-.(((((((....))))((-((.....)))----)........)))..------ ( -30.60, z-score = -1.18, R) >droMoj3.scaffold_6473 9832167 95 - 16943266 --GCCGUAGCUAAAAGGCAGUUAAAAACUGCGGCACGUAGUCAGUG-------GCAAU-GGGCGCGCGAAAGCGUUG-CACUGACUGCGAAUGAAUGAAAAGCCAG------ --(((((.(((....)))(((.....)))))))).(((((((((((-------((...-..))((((....))))..-))))))))))).................------ ( -39.20, z-score = -3.55, R) >consensus _GGUCUUGGU_AAAAGGCAGUUAAAAC_UGCGGCACGUAGUCGUUA_______GCGUUGAAAAGCGCGAAAGCGUAG_CAUUGACUGC___CAAAUGAAAAGCCAG______ ......((((......((((((((.......(((.....))).....................((((....)))).....)))))))).............))))....... (-18.58 = -18.97 + 0.39)

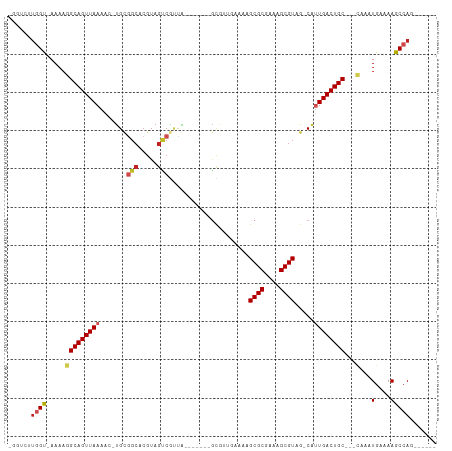
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:19 2011