| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,401,614 – 11,401,714 |
| Length | 100 |
| Max. P | 0.792658 |

| Location | 11,401,614 – 11,401,714 |
|---|---|
| Length | 100 |
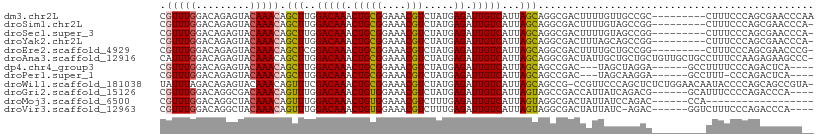
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Shannon entropy | 0.49422 |
| G+C content | 0.48256 |
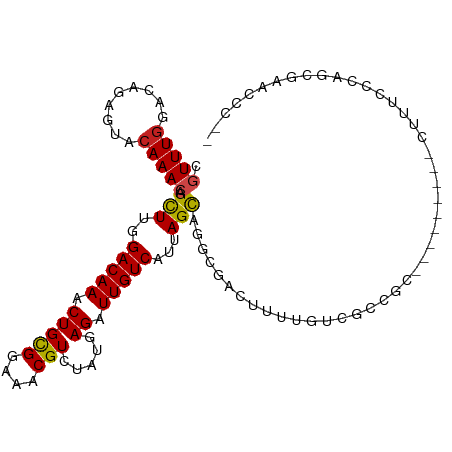
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -14.34 |
| Energy contribution | -13.99 |
| Covariance contribution | -0.35 |
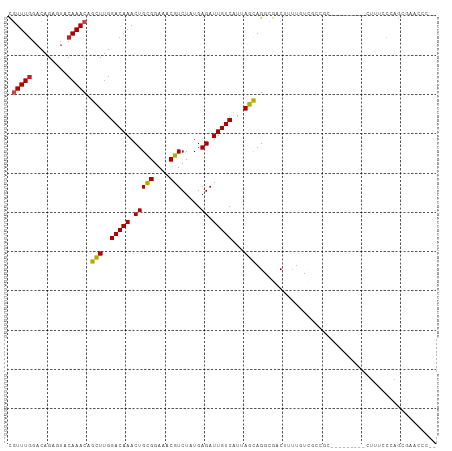
| Combinations/Pair | 1.17 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11401614 100 + 23011544 CGUUUGGACAGAGUACAAACAGCUUGGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGGCGACUUUUGUUGCCGC---------CUUUCCCAGCGAACCCAA ((((.(((.((.((.......(((..(((((.(((((....))).....)).)))))...))).((((((....))))))))---------)).))).))))....... ( -33.90, z-score = -3.09, R) >droSim1.chr2L 11210033 99 + 22036055 CGUUUGGACAGAGUACAAACAGCUUGGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGGCGACUUUUGUAGCCGG---------CUUUCCCAGCGAACCCA- ((((.(((..((((.......(((..(((((.(((((....))).....)).)))))...))).(((.((....)).))).)---------)))))).))))......- ( -29.90, z-score = -1.69, R) >droSec1.super_3 6796458 99 + 7220098 CGUUUGGACAGAGUACAAACAGCUUGGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGGCGACUUUUGUAGCCGG---------CUUUCCCAGCGAACCCA- ((((.(((..((((.......(((..(((((.(((((....))).....)).)))))...))).(((.((....)).))).)---------)))))).))))......- ( -29.90, z-score = -1.69, R) >droYak2.chr2L 7800066 99 + 22324452 CGUUUGGACAGAGUACAAACAGCUUGGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGGCGACUUUAGCAGCCGG---------CUUUCCCAGCGAACCCA- ((((.(((..((((.......(((..(((((.(((((....))).....)).)))))...))).(((..(....)..))).)---------)))))).))))......- ( -28.90, z-score = -1.38, R) >droEre2.scaffold_4929 12603096 99 - 26641161 CGUUUGGACAGAGUACAAACAGCUCGGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGGCGACUUUUGCUGCCGG---------CUUUCCCAGCGAACCCG- ((((.(((..((((.......(((..(((((.(((((....))).....)).)))))...))).((((.(....).)))).)---------)))))).))))......- ( -28.40, z-score = -0.79, R) >droAna3.scaffold_12916 12110165 108 - 16180835 CAUUUGGACAGAGUACAAACAGCUUGGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGGCGACUAUUGCUGCUGCUGUUGCUGCCUUUCCAAGAGAAGCCC- ..((((((.((.(((..((((((...(((((.(((((....))).....)).)))))...(((((.(((...))))))))))))))..))))).))))))........- ( -34.40, z-score = -1.97, R) >dp4.chr4_group3 6852110 96 + 11692001 CGUUUGGACAGAGUACAAACAGCUUGGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGCCGAC---UAGCUAGGA------GCCUUUUCCCAGACUCA---- .((((((..((((........(((..(((((.(((((....))).....)).)))))...)))(((....---..)))....------..))))..))))))...---- ( -25.60, z-score = -1.03, R) >droPer1.super_1 8300128 95 + 10282868 CGUUUGGACAGAGUACAAACAGCUUGGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGCCGAC---UAGCAAGGA------GCCUUU-CCCAGACUCA---- .((((((..((((........(((..(((((.(((((....))).....)).)))))...))).((....---..)).....------..))))-.))))))...---- ( -23.70, z-score = -0.51, R) >droWil1.scaffold_181038 335545 107 + 637489 UAUUUAGACAGAGUACAAACAGUUUCGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGCCG-CCGUUCCCAGCUCUCUGGAACAAUACCCCAGCAGCCGUA- ......(((((........((((((....))))))((....)).........)))))....((.((.(-(.(((((.((....))))))).........)).)).)).- ( -19.40, z-score = 0.57, R) >droGri2.scaffold_15126 7218920 99 - 8399593 CGUUUGGACAGGCGACAAACAGUUUGGACAAACUGUGGAAACGUCUAUGAGAUUGUCAUUAGUAGCCGACCAUUAUCAGACG------GCAUUUCCCCAGACCCA---- .((((((......((((((((((((....)))))))(....)..........))))).......((((.(........).))------))......))))))...---- ( -26.60, z-score = -1.48, R) >droMoj3.scaffold_6500 5059598 85 - 32352404 CGUUUGGACAGGCUACAAACAGUUUGGACAAACUGUGGAAACGUCUUUGAGAUUGUCAUUAGUAGGCGACUAUUAUCCAGAC------CCA------------------ .((((((((((.((.((((((((((....)))))))(....)....))))).)))))..((((((....))))))..)))))------...------------------ ( -25.10, z-score = -2.30, R) >droVir3.scaffold_12963 18076419 98 - 20206255 CGUUUGGACAGGCUACAAACAGUUUGGACAAACUGUGGAAACGUCUUUGAGAUUGUCAUUAGUAGGCGACUAUUAUC-AGAC------GGUCUUUCCCAGACCCA---- .(((((..((((((......))))))..)))))...(....)((((..(((((((((..((((((....))))))..-.)))------))))))....))))...---- ( -34.90, z-score = -3.77, R) >consensus CGUUUGGACAGAGUACAAACAGCUUGGACAAACUGCGGAAACGUCUAUGAGAUUGUCAUUAGCAGGCGACUUUUGUCGCCGC_________CUUUCCCAGCGAACCC__ .(((((.........))))).(((..(((((.(((((....))).....)).)))))...))).............................................. (-14.34 = -13.99 + -0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:29 2011