| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,626,831 – 14,626,941 |
| Length | 110 |
| Max. P | 0.995775 |

| Location | 14,626,831 – 14,626,941 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.68 |
| Shannon entropy | 0.49871 |
| G+C content | 0.50063 |
| Mean single sequence MFE | -17.75 |
| Consensus MFE | -13.13 |
| Energy contribution | -13.30 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995775 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
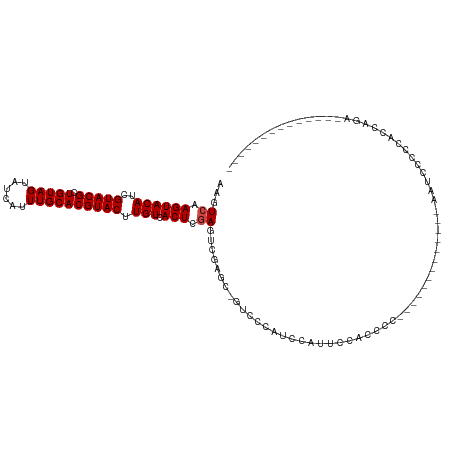
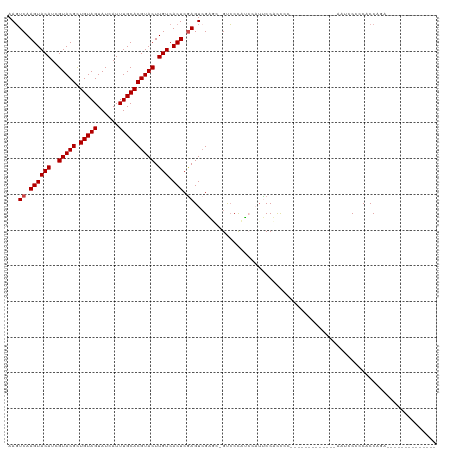
>dm3.chrX 14626831 110 + 22422827 AAGUCAAGUACAUCGUACGUUGUAGUAUCAUUUGCACGUACUUGUCACUCGAGUCGAGCCGUCCUUUCCAUUCCUUUCC----------CUUGAUUCCCCACCAGAAUCCUCUACCUUCA ...((.((((((..((((((.((((......)))))))))).))).))).((((((((.....................----------)))))))).......)).............. ( -18.20, z-score = -2.10, R) >droYak2.chrX 8853381 120 + 21770863 AAGUCAAGUACAUCGUACGUUGUAGUAUCAUUUGCACGUACUUGUCACUCGAGUCGAGCCGUCCUUUCCAUUCCUUUCCAUUCCUAUCCCUUGAAUCCCCACCAGAAAUACCCAUCCACA ...(((((.(((..((((((.((((......)))))))))).)))..((((...))))...............................))))).......................... ( -16.40, z-score = -1.63, R) >droSec1.super_32 79862 110 + 562834 AAGUCAAGUACAUCGUACGUUGUAGUAUCAUUUGCACGUACUUGUCACUCGAGUAGAGCCGUCCUUUCCAUUCCUUUCC----------CUCGAUUCCCCACCAGAAUCCUCCACCUCCA ..(..(((.((...((((((.((((......))))))))))......(((.....)))..)).)))..)..........----------...(((((.......)))))........... ( -17.40, z-score = -2.17, R) >dp4.chrXL_group1e 7868911 90 - 12523060 AAGUCAAGUACAUCGUACGCUGUAGUAUCAUUUGCACGUACUUGUCACUCGAGUCGU---GCCCCAAAGCUCCCACCGC-------------ACCCCCCCACCACC-------------- .......((((...))))((.(((((.......(((((.(((((.....))))))))---))......)))...)).))-------------..............-------------- ( -17.12, z-score = -2.23, R) >droPer1.super_17 334406 90 - 1930428 AAGUCAAGUACAUCGUACGCUGUAGUAUCAUUUGCACGUACUUGUCACUCGAGUCGU---GCCCCAAAGCUCCCACCAA-------------ACCCCCCCACCACC-------------- ......((((((..(((((.(((((......)))))))))).))).))).((((..(---(...))..)))).......-------------..............-------------- ( -15.30, z-score = -2.16, R) >droAna3.scaffold_12948 32785 90 - 692136 AAUUCAAGUACAUCGUACGCUGUAGUAUCAUUUGCACGUACUUGUCACUCAAGUCGGUG-GUGCCACCGAAUCCACC-C-------------ACUCCCCCUCCGA--------------- ......((((((..(((((.(((((......)))))))))).))).)))....((((((-....)))))).......-.-------------.............--------------- ( -22.10, z-score = -2.45, R) >consensus AAGUCAAGUACAUCGUACGCUGUAGUAUCAUUUGCACGUACUUGUCACUCGAGUCGAGC_GUCCCAUCCAUUCCACCCC_____________AAUCCCCCACCAGA______________ ...((.((((((..(((((.(((((......)))))))))).))).))).)).................................................................... (-13.13 = -13.30 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:17 2011