| Sequence ID | dm3.chrX |
|---|---|
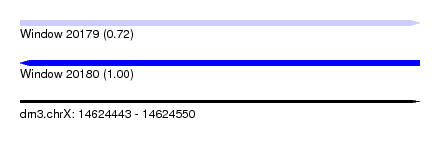
| Location | 14,624,443 – 14,624,550 |
| Length | 107 |
| Max. P | 0.997161 |

| Location | 14,624,443 – 14,624,550 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.29 |
| Shannon entropy | 0.58393 |
| G+C content | 0.56554 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -17.52 |
| Energy contribution | -20.22 |
| Covariance contribution | 2.70 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.722085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
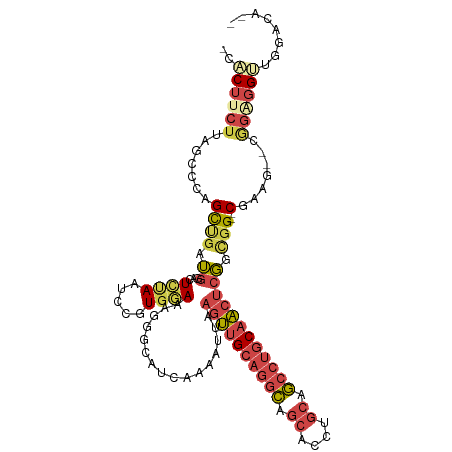
>dm3.chrX 14624443 107 + 22422827 -CACUUCUUAGCCCAGCUGAUGGACUUUAAUUCGUGAAAAGGGCAUCAAAAUUAAGUUGCAGGCAGCACCUGCAGCCUGCAACUCGGCGG--CGAAG--UGGAGGUUGGACA-- -((((((...(((....(((((..((((.........))))..)))))......((((((((((.((....)).)))))))))).)))..--.))))--))...........-- ( -40.30, z-score = -2.35, R) >droAna3.scaffold_12948 28057 112 - 692136 CUGCACCUGCGGCCAGUGAUGAGAAUGGAAAUGGU--AAGGGAAAUGUGGCAGGGACUGGAGAGGCAGCCUGCUUGCUGCAGCAGCGAGGAACGAGACGCUGCGGCCAAGAACA ..((....))((((.....................--........((..((((((.(((......))))))...)))..))((((((..........))))))))))....... ( -34.20, z-score = 0.47, R) >droEre2.scaffold_4690 6385972 109 - 18748788 -CCCUUCUUAGCGCAGCCGAUGGACUCCAGUCCGUGGAAGGGGCAUCCACAUUAAGUUGCAGGCAGCACCUGCAGCCCGCAACUCGGCGG--CGCAGCGCGGGGGUUGGACA-- -((((.(...((((.(((((.((((....))))(((((.......))))).....(((((.(((.((....)).))).)))))))))).)--)))...).))))........-- ( -49.30, z-score = -1.61, R) >droYak2.chrX 8851041 101 + 21770863 -CACUACUUAGAGCAGCUGUUGGACUCCAAUCCGUGGAGAGGGCAUCAAAAUUAAGUUGCAUGCAGCACCUGCAGCCUGCCACUCGGUGG--CGGAG-ACAAGGG--------- -...........((((.(((((..(((((.....)))))...(((.(........).)))...))))).))))..((((((((...))))--)(...-.).))).--------- ( -32.50, z-score = -0.18, R) >droSec1.super_32 77820 107 + 562834 -CACUUCUUAGCCCAGCUGAUGGACUCUAAUCCCUGGAAAGGGCAUCAAAAUUAAGUUGCAGGCAGCACCUGCAGCCUGCAACUCGUCGG--CGAAG--UGGAGGUUGGACA-- -.((((((..((...(((((((.........((((....))))...........((((((((((.((....)).))))))))))))))))--)...)--)))))))......-- ( -42.50, z-score = -2.61, R) >droSim1.chrX 11284218 107 + 17042790 -CACUUCUCAGCCCAGCUGAUGGACUCUAAUCCCUGGAAAGGGCAUCAAAAUUAAGUUGCAGGCAGCACCUGCAGCCUGCAACUCGGCGG--CGAAG--UGGAGGUUGGACA-- -((((((.(.(((....(((((.........((((....)))))))))......((((((((((.((....)).)))))))))).))).)--.))))--))...........-- ( -43.10, z-score = -2.28, R) >consensus _CACUUCUUAGCCCAGCUGAUGGACUCUAAUCCGUGGAAAGGGCAUCAAAAUUAAGUUGCAGGCAGCACCUGCAGCCUGCAACUCGGCGG__CGAAG__CGGAGGUUGGACA__ ..((((((..((((.......(((......))).......))))..........((((((((((.((....)).))))))))))................))))))........ (-17.52 = -20.22 + 2.70)

| Location | 14,624,443 – 14,624,550 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.29 |
| Shannon entropy | 0.58393 |
| G+C content | 0.56554 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -21.38 |
| Energy contribution | -24.16 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.997161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
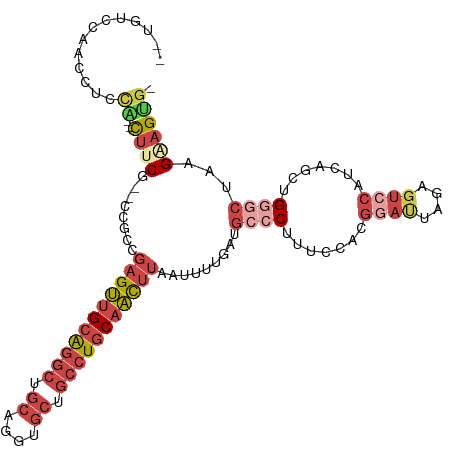
>dm3.chrX 14624443 107 - 22422827 --UGUCCAACCUCCA--CUUCG--CCGCCGAGUUGCAGGCUGCAGGUGCUGCCUGCAACUUAAUUUUGAUGCCCUUUUCACGAAUUAAAGUCCAUCAGCUGGGCUAAGAAGUG- --...........((--((((.--..((((((((((((((.((....)).)))))))))))....((((((..((((.........))))..))))))...)))...))))))- ( -42.30, z-score = -4.40, R) >droAna3.scaffold_12948 28057 112 + 692136 UGUUCUUGGCCGCAGCGUCUCGUUCCUCGCUGCUGCAGCAAGCAGGCUGCCUCUCCAGUCCCUGCCACAUUUCCCUU--ACCAUUUCCAUUCUCAUCACUGGCCGCAGGUGCAG (((((((((((((((((..........))))))........((((((((......)))..)))))............--.....................))))).))).))). ( -29.70, z-score = -0.53, R) >droEre2.scaffold_4690 6385972 109 + 18748788 --UGUCCAACCCCCGCGCUGCG--CCGCCGAGUUGCGGGCUGCAGGUGCUGCCUGCAACUUAAUGUGGAUGCCCCUUCCACGGACUGGAGUCCAUCGGCUGCGCUAAGAAGGG- --........(((..(...(((--(.((((((((((((((.((....)).))))))))).....(((((.......)))))((((....)))).))))).))))...)..)))- ( -54.20, z-score = -3.51, R) >droYak2.chrX 8851041 101 - 21770863 ---------CCCUUGU-CUCCG--CCACCGAGUGGCAGGCUGCAGGUGCUGCAUGCAACUUAAUUUUGAUGCCCUCUCCACGGAUUGGAGUCCAACAGCUGCUCUAAGUAGUG- ---------.....(.-((((.--((...(((.((((.(((((((...))))).))..(........).))))..)))...))...)))).).....(((((.....))))).- ( -27.20, z-score = 0.86, R) >droSec1.super_32 77820 107 - 562834 --UGUCCAACCUCCA--CUUCG--CCGACGAGUUGCAGGCUGCAGGUGCUGCCUGCAACUUAAUUUUGAUGCCCUUUCCAGGGAUUAGAGUCCAUCAGCUGGGCUAAGAAGUG- --...........((--((((.--.....(((((((((((.((....)).)))))))))))...((((((.((((....))))))))))(((((.....)))))...))))))- ( -42.60, z-score = -3.16, R) >droSim1.chrX 11284218 107 - 17042790 --UGUCCAACCUCCA--CUUCG--CCGCCGAGUUGCAGGCUGCAGGUGCUGCCUGCAACUUAAUUUUGAUGCCCUUUCCAGGGAUUAGAGUCCAUCAGCUGGGCUGAGAAGUG- --...........((--((((.--(.((((((((((((((.((....)).))))))))))).((((((((.((((....))))))))))))..........))).).))))))- ( -45.10, z-score = -3.35, R) >consensus __UGUCCAACCUCCA__CUUCG__CCGCCGAGUUGCAGGCUGCAGGUGCUGCCUGCAACUUAAUUUUGAUGCCCUUUCCACGGAUUAGAGUCCAUCAGCUGGGCUAAGAAGUG_ .................((((........(((((((((((.((....)).))))))))))).........((((.......((((....)))).......))))...))))... (-21.38 = -24.16 + 2.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:17 2011