| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,604,582 – 14,604,672 |
| Length | 90 |
| Max. P | 0.731458 |

| Location | 14,604,582 – 14,604,672 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 53.80 |
| Shannon entropy | 0.90171 |
| G+C content | 0.42247 |
| Mean single sequence MFE | -20.69 |
| Consensus MFE | -8.37 |
| Energy contribution | -8.25 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.94 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731458 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
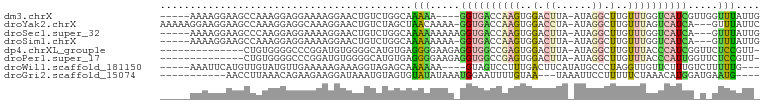
>dm3.chrX 14604582 90 + 22422827 -----AAAAGGAAGCCAAAGGAGGAAAAGGAACUGUCUGGCAAAAA----GGUGACCAAGUGGACUUA-AUAGGCUUGUUUGGUCAUCGUUGGUUUAUUG -----......(((((((((..((........))..))........----((((((((((..(.((..-...)).)..)))))))))).))))))).... ( -23.80, z-score = -1.83, R) >droYak2.chrX 8832887 95 + 21770863 AAAAAGGAAGGAAGCCAAAGGAGGCAAAGGAACUGUCUAGCUAACAAAA-GGUGACCAAGUGGACCUA-AUAGGCUUGUUUAGUCAUCA---GUUUAUUC .............(((......)))....(((((((.......))....-((((((.(((..(.((..-...)).)..))).)))))))---)))).... ( -18.60, z-score = 0.29, R) >droSec1.super_32 57967 91 + 562834 -----AAAAGGAAGCCCAAGGAGGAAAAGGAACUGUCUGGCAAAAAAAAAGGUGACCAAGUGGACUUA-AUAGGCUUGUUUGGUCAUCA---GUUUAUUG -----........(((..((..((........))..))))).........((((((((((..(.((..-...)).)..)))))))))).---........ ( -21.80, z-score = -1.04, R) >droSim1.chrX 11264528 90 + 17042790 -----AAAAGGAAGCCAAAGGAGGAAAAGGAACUGUCUGGCAAAAAAAA-GGUGACCAAGUGGACUUA-AUAGGCUUGUUUGGUCAUCA---GUUUAUUG -----........((((.((............))...))))........-((((((((((..(.((..-...)).)..)))))))))).---........ ( -22.40, z-score = -1.73, R) >dp4.chrXL_group1e 7844831 84 - 12523060 --------------CUGUGGGGCCCGGAUGUGGGGCAUGUGAGGGGAAGAGGUGGCCGAGUGGACUUA-AUAGGCUUGUUUACCCAUCGGUUCUCCGUU- --------------.......((((.......))))......((((((..(((((....((((((...-........)))))))))))..))))))...- ( -26.50, z-score = 0.53, R) >droPer1.super_17 310567 84 - 1930428 --------------CUGUGGGGCCCGGAUGUGGGGCAUGUGAGGGGAAGAGGUGGCCGAGUGGACUUA-AUAGGCUUGUUUACCCAUUGGUUCUCCGUU- --------------.......((((.......))))......((((((..(((((....((((((...-........)))))))))))..))))))...- ( -24.80, z-score = 0.78, R) >droWil1.scaffold_181150 759822 88 - 4952429 -----AAAUUCAUGUUGUAUGUUGAAAAAGAAAGGUAGAGCAAAAAA----GUAGUCCUUUGACUUCAUAUGCCCUAGGUUGUUCUUUGUCUUUUUG--- -----...................(((((((.....((((((.....----(.((((....)))).)....(((...)))))))))...))))))).--- ( -14.00, z-score = -0.32, R) >droGri2.scaffold_15074 3457431 82 - 7742996 -----------AACCUUAAACAGAAGAAGGAUAAAUGUAGUGUAUAUAAAUGGAAUUUUGUAA---UAAAUUCCUUUUUCUAAACAUGGAUGAAUG---- -----------..((......((((((((((.........(((.((((((......)))))))---))...))))))))))......)).......---- ( -13.60, z-score = -1.31, R) >consensus _____AAAAGGAAGCCAAAGGAGGAAAAGGAACUGUCUGGCAAAAAAAA_GGUGACCAAGUGGACUUA_AUAGGCUUGUUUAGUCAUCG_U_GUUUAUU_ ..........................................(((.....((((((((((..(.(((....))).)..)))))))))).....))).... ( -8.37 = -8.25 + -0.12)

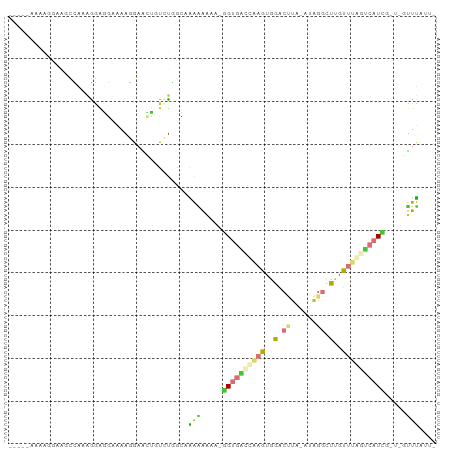
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:13 2011