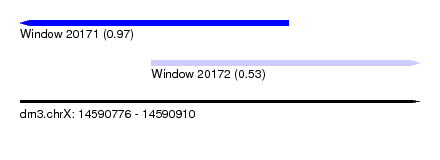
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,590,776 – 14,590,910 |
| Length | 134 |
| Max. P | 0.970997 |

| Location | 14,590,776 – 14,590,866 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 64.88 |
| Shannon entropy | 0.59955 |
| G+C content | 0.45353 |
| Mean single sequence MFE | -17.81 |
| Consensus MFE | -11.14 |
| Energy contribution | -11.58 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14590776 90 - 22422827 ---CGCUCACUUU-CGGCAACAUUGUUCCCCGGAGAAUUCCACC--AUUCUUCGGCAGAUACAAAAA----AAAGAA------ACAC-AGAUACAGAUACAGUCUGG ---.......(((-((....).((((((.(((((((((......--)))))))))..)).))))...----...)))------)..(-((((.........))))). ( -19.60, z-score = -2.28, R) >droSim1.chrX 11249053 100 - 17042790 ---CGCUCACUUU-CGGCAACAUUGUUCCCCGGAGAAUUCCUCC--AUUCUUCGGCAGAUACAAAAACCAUAUACACGUACAGAUAC-AGAUACAGAUACAGUCUGG ---.(((......-.)))....((((((.(((((((((......--)))))))))..)).))))......................(-((((.........))))). ( -18.40, z-score = -1.18, R) >droSec1.super_32 42879 94 - 562834 ---CGCUCACUUU-CGGCAACAUUGUUCCCCGGAGAAUUCCUCC--AUUCUUCGGCAGAUACAAAAACCAUAUACAC------GUAC-AGAUACAGAUACAGUCUGG ---.(((......-.)))....((((((.(((((((((......--)))))))))..)).)))).............------...(-((((.........))))). ( -18.40, z-score = -1.50, R) >droYak2.chrX 8820310 94 - 21770863 ---CGCUGACUUU-CGGCAACAUUGUUCCCCGGAGAAUUCCUCC--AUUCUUCGGCAGAUACAAUAACC-UAUACAG------AUACAAGAUACAGAUGCAGUCUGG ---.((((.....-))))...(((((((.(((((((((......--)))))))))..)).)))))....-.......------..........(((((...))))). ( -22.80, z-score = -1.96, R) >droWil1.scaffold_181150 743678 72 + 4952429 UGCUGCCUGCUCCACAGCAUCUUUGGCCCACAAAUCAUUUUUGCUAAACAUUCAACACUCUUAACAGGCUGG----------------------------------- ....(((((((....)))...((((((...............)))))).................))))...----------------------------------- ( -9.86, z-score = 0.76, R) >consensus ___CGCUCACUUU_CGGCAACAUUGUUCCCCGGAGAAUUCCUCC__AUUCUUCGGCAGAUACAAAAACC_UAUACAC______AUAC_AGAUACAGAUACAGUCUGG ....(((........)))....((((...(((((((((........))))))))).....)))).............................((((.....)))). (-11.14 = -11.58 + 0.44)

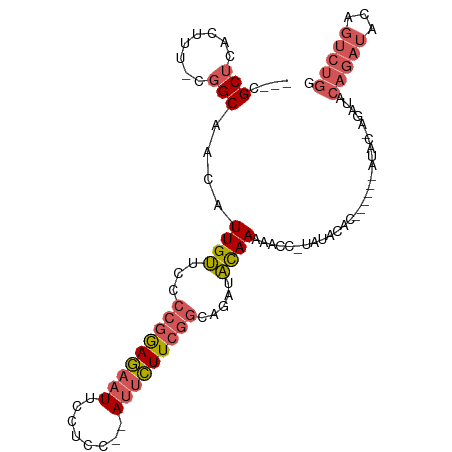
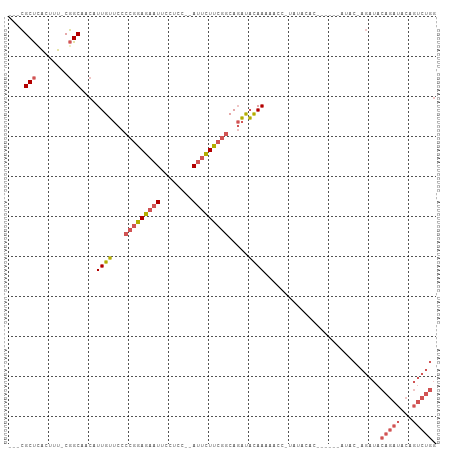
| Location | 14,590,820 – 14,590,910 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 76.07 |
| Shannon entropy | 0.43753 |
| G+C content | 0.42055 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -9.81 |
| Energy contribution | -11.26 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.526791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14590820 90 + 22422827 AAGAAUGGUGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGGAAAG ....((((((((....))).((((((...(((..(....)..)))(..(((.......)))..)..))))))....)))))......... ( -21.70, z-score = -1.21, R) >droSim1.chrX 11249107 90 + 17042790 AAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGAAAAG ..((.(((((.....)))))((((((...(((..(....)..)))(..(((.......)))..)..)))))).....))........... ( -22.60, z-score = -2.09, R) >droSec1.super_32 42927 90 + 562834 AAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGAAAAG ..((.(((((.....)))))((((((...(((..(....)..)))(..(((.......)))..)..)))))).....))........... ( -22.60, z-score = -2.09, R) >droYak2.chrX 8820358 90 + 21770863 AAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUCAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGAAAAG ..((.(((((.....)))))((((((...((((.(....).))))(..(((.......)))..)..)))))).....))........... ( -25.10, z-score = -2.85, R) >droEre2.scaffold_4690 6356872 89 - 18748788 AAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGGCAAGCACGUUCCCAUAAAUCAUAAAGAAGG- ..((.(((((.....)))))((((((..(((((((....((...(.....)..))..))).)))).)))))).....))..........- ( -21.70, z-score = -1.51, R) >dp4.chrXL_group1e 7832077 72 - 12523060 -----------------AGAGUGAAUGA-GUGGCACAAUGUUAGCAAAUGGAAAUGAGCCACAAGAGUUCCUAUAAAUUACAAGUACAAG -----------------..((.((((..-(((((.((...(((.....)))...)).)))))....)))))).................. ( -11.70, z-score = -0.75, R) >droPer1.super_17 297720 72 - 1930428 -----------------AGAGUGAAUGA-GUGGCACAAUGUUAGCAAAUGGAAAUGAGCCACAAGAGUUCCUAUAAAUUACAAGUACAAG -----------------..((.((((..-(((((.((...(((.....)))...)).)))))....)))))).................. ( -11.70, z-score = -0.75, R) >consensus AAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGAAAAG ......((((.....)))).((((((...(((..(....)..)))...(((.......))).....)))))).................. ( -9.81 = -11.26 + 1.45)
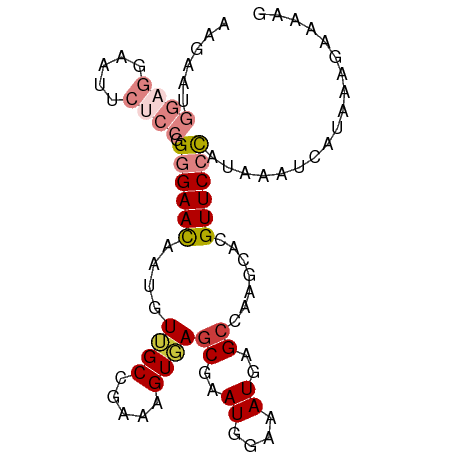
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:10 2011