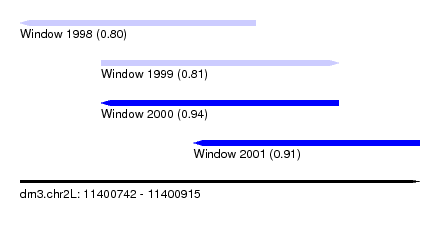
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,400,742 – 11,400,915 |
| Length | 173 |
| Max. P | 0.941190 |

| Location | 11,400,742 – 11,400,844 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 59.86 |
| Shannon entropy | 0.69571 |
| G+C content | 0.46552 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -11.24 |
| Energy contribution | -12.80 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
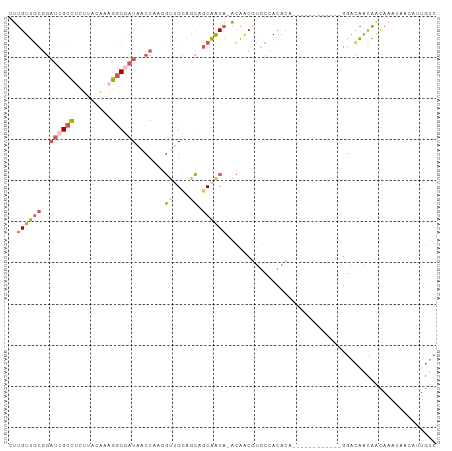
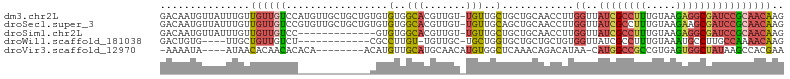
>dm3.chr2L 11400742 102 - 23011544 GUGUGGCACGUUGU-UGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCGCAACAAGUGCCGAAGACGCACGAUACCUAAAAGGUAUUUAAA----- (((((((((.((((-((((((....))).....((..((((((((.....))))))))))))))))))))))....))))..((((((.....))))))....----- ( -39.30, z-score = -3.73, R) >droSec1.super_3 6795608 102 - 7220098 GUGUGGCACGUUGU-UGUUGCAGCUGCAACCUUGGUUAUCGCCUUUGUAAGAAGCGAUCCGCAACAAGUGCCGAAGACGCACGAUACAUAAAAGUUAUUUAAA----- (((((((((.((((-((((((....))).....((..(((((.(((....))))))))))))))))))))))....)))).......................----- ( -27.90, z-score = -0.71, R) >droSim1.chr2L 11209158 98 - 22036055 ----GGCACGUUGU-UGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCGCAACAAGUGCCGAAGACGCACGAUACCUAAAAGUUAUUUAAA----- ----(((((.((((-((((((....))).....((..((((((((.....))))))))))))))))))))))...............................----- ( -32.10, z-score = -2.46, R) >droWil1.scaffold_181038 334868 89 - 637489 ------------------UGGUGCUGCUGCUGUGGUUAUCGCCUUUGUAAAUGCCUUGCCAAAACAAGUGUCGAAUGCACAUAAUGGGUACAA-UUACUUAUUGGCCA ------------------.(((((..(....)..).))))(((..((((...((((((......)))).))....))))...((((((((...-.))))))))))).. ( -18.10, z-score = 1.68, R) >droVir3.scaffold_12970 6029902 88 - 11907090 -------AUGCAACAUGUGGCUCAAACAGACAUAA-CAUGGCCGCCGUGAGUGGCUAUAAGCCACGAACAGGGGUCGCUCACCACAUCUAUAUAUA------------ -------.......((((((.((.....)).....-...((((.((((..(((((.....)))))..)).)))))).....)))))).........------------ ( -25.10, z-score = -1.17, R) >consensus ____GGCACGUUGU_UGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCGCAACAAGUGCCGAAGACGCACGAUACCUAAAAGUUAUUUAAA_____ ...............((((((......(((....)))((((((((.....))))))))..)))))).((((.......)))).......................... (-11.24 = -12.80 + 1.56)

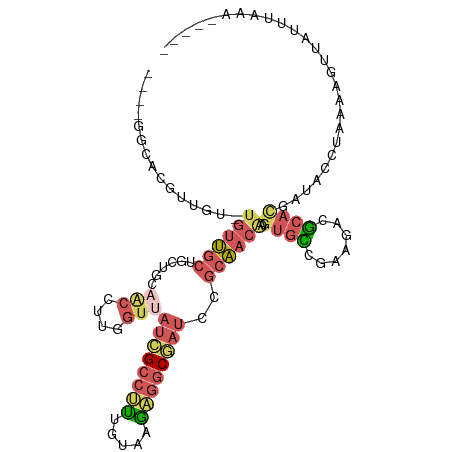
| Location | 11,400,777 – 11,400,880 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 61.48 |
| Shannon entropy | 0.65631 |
| G+C content | 0.47251 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -8.76 |
| Energy contribution | -9.28 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810367 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 11400777 103 + 23011544 CUUGUUGCGGAUCGCCUCUUACAAAGGCGAUAACCAAGGUUGCAGCAGCAACA-ACAACGUGCCACACACAGCAGCAACAUGGACAACAACAAAUAACAUUGUC .((((((((((((((((.......)))))))..))...(((((....))))).-.....(((.....))).)))))))....(((((............))))) ( -28.60, z-score = -2.53, R) >droSec1.super_3 6795643 103 + 7220098 CUUGUUGCGGAUCGCUUCUUACAAAGGCGAUAACCAAGGUUGCAGCUGCAACA-ACAACGUGCCACACACAGCAGCAACACGGACAACAACAAAUAACAUUGUC .((((((((((((((((.......)))))))..))...(((((....))))).-.....(((.....))).)))))))....(((((............))))) ( -26.20, z-score = -1.65, R) >droSim1.chr2L 11209193 90 + 22036055 CUUGUUGCGGAUCGCCUCUUACAAAGGCGAUAACCAAGGUUGCAGCAGCAACA-ACAACGUGCCACAC-------------GGACAACAACAAAUAACAUUGUC .((((((..((((((((.......)))))))..((...(((((....))))).-.....(((...)))-------------)).)..))))))........... ( -25.00, z-score = -2.43, R) >droWil1.scaffold_181038 334907 86 + 637489 CUUGUUUUGGCAAGGCAUUUACAAAGGCGAUAACCACAGCAGCAGCACCAGCA-GCAACA-ACAAGGCG------------AGACAACAGCAA----CACAGUC (((((((((.....((.........((......))...((....)).......-))....-.)))))))------------))..........----....... ( -15.00, z-score = 0.02, R) >droVir3.scaffold_12970 6029930 90 + 11907090 UUCGUGGCUUAUAGCCACUCACGGCGGCCAUG-UUAUGUCUGUUUGAGCCACAUGUUGCAUGCAACAUGU--------UGUGUGUUGUGUUAU----UAUUUU- ...(((((.....))))).(((((((((((..-...........)).)))((((((((....))))))))--------.....))))))....----......- ( -29.12, z-score = -1.84, R) >consensus CUUGUUGCGGAUCGCCUCUUACAAAGGCGAUAACCAAGGUUGCAGCAGCAACA_ACAACGUGCCACACA____________GGACAACAACAAAUAACAUUGUC ...(((((..((((((.........)))))).......((....)).))))).................................................... ( -8.76 = -9.28 + 0.52)

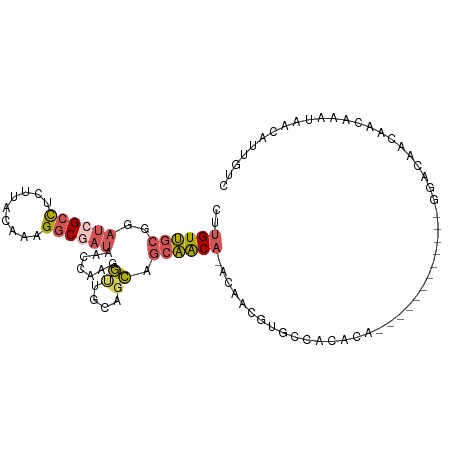
| Location | 11,400,777 – 11,400,880 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 61.48 |
| Shannon entropy | 0.65631 |
| G+C content | 0.47251 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -12.14 |
| Energy contribution | -12.86 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941190 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
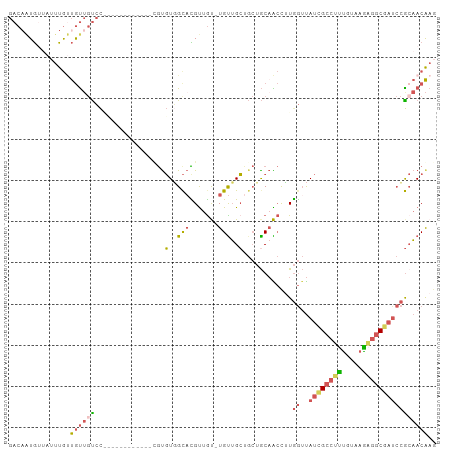
>dm3.chr2L 11400777 103 - 23011544 GACAAUGUUAUUUGUUGUUGUCCAUGUUGCUGCUGUGUGUGGCACGUUGU-UGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCGCAACAAG (((((((........)))))))..(((((((((.(((.(..(((......-)))..)))).))).....((..((((((((.....)))))))))))))))).. ( -35.20, z-score = -2.44, R) >droSec1.super_3 6795643 103 - 7220098 GACAAUGUUAUUUGUUGUUGUCCGUGUUGCUGCUGUGUGUGGCACGUUGU-UGUUGCAGCUGCAACCUUGGUUAUCGCCUUUGUAAGAAGCGAUCCGCAACAAG (((((((........))))))).((((..(........)..))))(((((-.(((((....)))))...((..(((((.(((....)))))))))))))))... ( -31.40, z-score = -1.18, R) >droSim1.chr2L 11209193 90 - 22036055 GACAAUGUUAUUUGUUGUUGUCC-------------GUGUGGCACGUUGU-UGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCGCAACAAG (((((((........))))))).-------------(((...)))(((((-.(((((....)))))...((..((((((((.....)))))))))))))))... ( -29.10, z-score = -2.05, R) >droWil1.scaffold_181038 334907 86 - 637489 GACUGUG----UUGCUGUUGUCU------------CGCCUUGU-UGUUGC-UGCUGGUGCUGCUGCUGUGGUUAUCGCCUUUGUAAAUGCCUUGCCAAAACAAG ....(((----..((....))..------------)))(((((-(.(((.-.((.((.((...(((...(((....)))...)))...)))).))))))))))) ( -17.10, z-score = 0.62, R) >droVir3.scaffold_12970 6029930 90 - 11907090 -AAAAUA----AUAACACAACACACA--------ACAUGUUGCAUGCAACAUGUGGCUCAAACAGACAUAA-CAUGGCCGCCGUGAGUGGCUAUAAGCCACGAA -......----...........(((.--------((((((((....))))))))(((.((...........-..)))))...))).(((((.....)))))... ( -25.82, z-score = -2.66, R) >consensus GACAAUGUUAUUUGUUGUUGUCC____________CGUGUGGCACGUUGU_UGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCGCAACAAG ...............((((((.................(..(((.......)))..)............((..((((((((.....)))))))))))))))).. (-12.14 = -12.86 + 0.72)
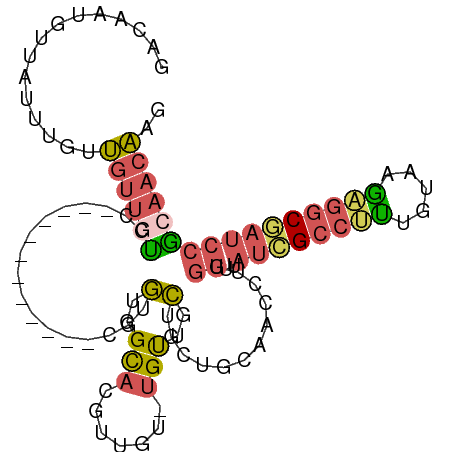
| Location | 11,400,817 – 11,400,915 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 65.15 |
| Shannon entropy | 0.54465 |
| G+C content | 0.45380 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -17.48 |
| Energy contribution | -21.48 |
| Covariance contribution | 4.00 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913152 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 11400817 98 - 23011544 ----GCAGUUGCUCGUUGCAGGAACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCAUGUUGCUGCUGUGUGUGGCACGUUGUUGUUGCUGCUGCA ----((((((((.....))))...((((((((((.....(((((((........))))))).((((.((..(.....)..)))))).)))))))))))))). ( -36.60, z-score = -1.54, R) >droSec1.super_3 6795683 98 - 7220098 ----GCAGUUGCUCGCUGCAGGAACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUGCUGCUGUGUGUGGCACGUUGUUGUUGCAGCUGCA ----((((((((..((.((((((.(((((((((.(..((.....))..).))))))))))))((((..(........)..))))..))).)).)))))))). ( -41.40, z-score = -2.44, R) >droSim1.chr2L 11209233 85 - 22036055 ----GCAGUUGCUCGCUGCAGGAACAGUAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCC-------------GUGUGGCACGUUGUUGUUGCUGCUGCA ----((((((((.....))))...((((((((((.....(((((((........)))))))(-------------(((...))))..)))))))))))))). ( -29.10, z-score = -1.21, R) >droVir3.scaffold_12970 6029963 96 - 11907090 UCAAUUGGAGGCGCACUGCAACAUAAGCAACAAUUGUCAAAAAUAAUAACACAACACACAACAUGUUGC------AUGCAACAUGUGGCUCAAACAGACAUA .......(((.((((.((((......((((((.((((....................))))..))))))------.))))...)))).)))........... ( -17.55, z-score = -0.41, R) >consensus ____GCAGUUGCUCGCUGCAGGAACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCAUGUUGC______GUGUGGCACGUUGUUGUUGCAGCUGCA ..............((.((((.(((((((((........(((((((........))))))).(((((((........)))))))))))))))).)))).)). (-17.48 = -21.48 + 4.00)

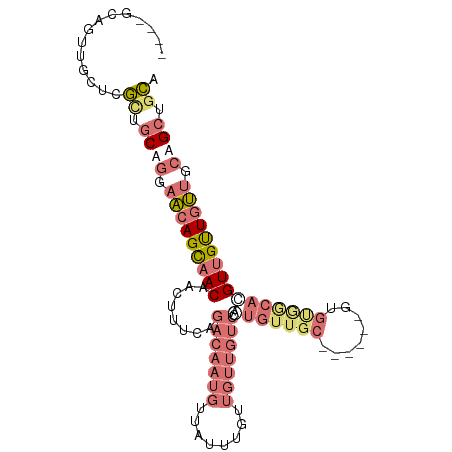
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:28 2011