| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,577,092 – 14,577,183 |
| Length | 91 |
| Max. P | 0.799121 |

| Location | 14,577,092 – 14,577,183 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 85.50 |
| Shannon entropy | 0.28113 |
| G+C content | 0.39691 |
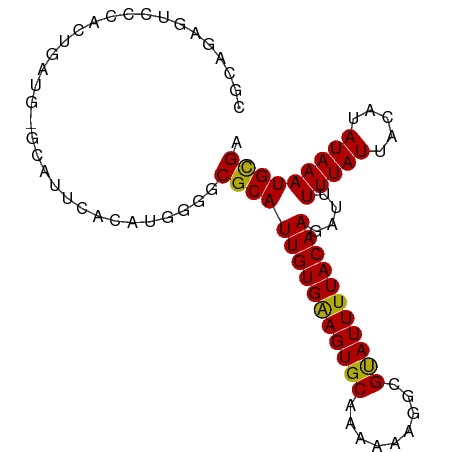
| Mean single sequence MFE | -23.44 |
| Consensus MFE | -15.36 |
| Energy contribution | -14.92 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
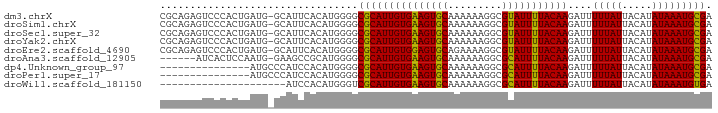
>dm3.chrX 14577092 91 - 22422827 CGCAGAGUCCCACUGAUG-GCAUUCACAUGGGGCGCAUUGUGAAGUGCAAAAAAGGCGUAUUUUACAAGAUUUUUAUUACAUAUAAAUGCGA (((((.((((((.(((..-....)))..)))))).).(((((((((((.........)))))))))))...................)))). ( -25.80, z-score = -2.33, R) >droSim1.chrX 11235443 91 - 17042790 CGCAGAGUCCCACUGAUG-GCAUUCACAUGGGGCGCAUUGUGAAGUGCAAAAAAGGCGUAUUUUACAAGAUUUUUAUUACAUAUAAAUGCGA (((((.((((((.(((..-....)))..)))))).).(((((((((((.........)))))))))))...................)))). ( -25.80, z-score = -2.33, R) >droSec1.super_32 29352 91 - 562834 CGCAGAGUCCCACUGAUG-GCAUUCACAUGGGGCGCAUUGUGAAGUGCAAAAAAGGCGUAUUUUACAAGAUUUUUAUUACAUAUAAAUGCGA (((((.((((((.(((..-....)))..)))))).).(((((((((((.........)))))))))))...................)))). ( -25.80, z-score = -2.33, R) >droYak2.chrX 8806092 91 - 21770863 CGCAGAGUCCCACUGAUG-GCAUUCACAUGGGGCGCAUUGUGAAGUGCAAAAAAGGCGUAUUUUACAAGAUUUUUAUUACAUAUAAAUGCGA (((((.((((((.(((..-....)))..)))))).).(((((((((((.........)))))))))))...................)))). ( -25.80, z-score = -2.33, R) >droEre2.scaffold_4690 6342747 91 + 18748788 CGCAGAGUCCCACUGAUG-GCAUUCACAUGGGGCGCAUUGUGGAGUGCAGAAAAGGCGUAUUUUACAAGAUUUUUAUUACAUAUAAAUGCGA (((((.((((((.(((..-....)))..)))))).).(((((((((((.........)))))))))))...................)))). ( -25.40, z-score = -1.99, R) >droAna3.scaffold_12905 526276 85 + 569613 ------AUCACUCCAAUG-GAAGCCGCAUGGGGCGCAUUGUGAAGUGCAAAAAAGGCGCAUUUUACAAGAUUUUUAUUACAUAUAAAUGCGA ------....(((((.((-(...)))..)))))(((((((((((((((.........)))))))))))....(((((.....))))))))). ( -21.60, z-score = -1.28, R) >dp4.Unknown_group_97 48557 77 - 51274 ---------------AUGCCCAUCCACAUGGGGCGCAUUGUGAAGUGCAAAAAAGGCGCAUUUUACAAGAUUUUUAUUACAUAUAAAUGCGA ---------------...(((((....))))).(((((((((((((((.........)))))))))))....(((((.....))))))))). ( -22.10, z-score = -2.21, R) >droPer1.super_17 282274 77 + 1930428 ---------------AUGCCCAUCCACAUGGGGCGCAUUGUGAAGUGCAAAAAAGGCGCAUUUUACAAGAUUUUUAUUACAUAUAAAUGCGA ---------------...(((((....))))).(((((((((((((((.........)))))))))))....(((((.....))))))))). ( -22.10, z-score = -2.21, R) >droWil1.scaffold_181150 728225 71 + 4952429 ---------------------AUCCACAUGGGUCGCAUUGUGAAGUGCAAAAAAGGCGCAUUUUACAAGAUUUUUAUUACAUAUAAAUGUGA ---------------------.(((....)))((((((((((((((((.........)))))))))))....(((((.....)))))))))) ( -16.60, z-score = -1.85, R) >consensus CGCAGAGUCCCACUGAUG_GCAUUCACAUGGGGCGCAUUGUGAAGUGCAAAAAAGGCGUAUUUUACAAGAUUUUUAUUACAUAUAAAUGCGA .................................(((((((((((((((.........)))))))))))....(((((.....))))))))). (-15.36 = -14.92 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:07 2011