| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,568,319 – 14,568,418 |
| Length | 99 |
| Max. P | 0.508521 |

| Location | 14,568,319 – 14,568,418 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.15 |
| Shannon entropy | 0.59282 |
| G+C content | 0.54486 |
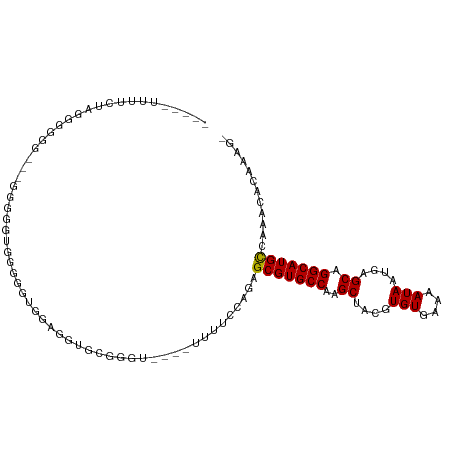
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -13.35 |
| Energy contribution | -14.73 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
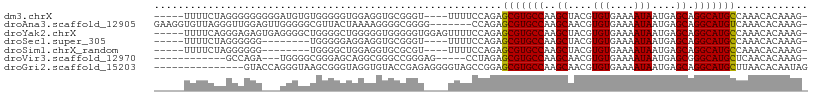
>dm3.chrX 14568319 99 + 22422827 -----UUUUCUAGGGGGGGGGAUGUGUGGGGGUGGAGGUGCGGGU----UUUUCCAGAGCGUGCCAAGCUACGUGUGAAAAUAAUGAGCAGGCAUGCCAAACACAAAG- -----((..((.....))..))(((((..(..((((((.......----.))))))..(((((((..(((...(((....)))...))).))))))))..)))))...- ( -30.70, z-score = -1.44, R) >droAna3.scaffold_12905 514054 101 - 569613 GAAGGUGUUAGGGUUGGAGUUGGGGGCGUUACUAAAAGGGGCGGGG-------CCAGAGCGUGCCAAGCAACGUGUGAAAAUAAUGAGCAGGCAUGUCAAACACAAAG- ....(((((...............(((.(..((......))..).)-------)).((.((((((..((....(((....)))....)).)))))))).)))))....- ( -25.70, z-score = -0.86, R) >droYak2.chrX 8797142 103 + 21770863 -----UUUUCAGGGAGAGUGAGGGGCUGGGGCUGGGGGUGGGGGUGGAGUUUUCCAGAGCGUGCCAAGCUACGUGUGAAAAUAAUGAGCAGGCAUGCCAAACACAAAG- -----.(..(((....(((.....)))....)))..)(((....(((......)))(.(((((((..(((...(((....)))...))).))))))))...)))....- ( -28.30, z-score = 0.04, R) >droSec1.super_305 15894 91 + 17228 -----UUUUCUAGGGGGG--------UGGGGGAGGAGGUGCGGGU----UUUUCCAGAGCGUGCCAAGCUACGUGUGAAAAUAAUGAGCAGGCAUGCCAAACACAAAG- -----............(--------((..(((((((.......)----)))))).(.(((((((..(((...(((....)))...))).))))))))...)))....- ( -26.00, z-score = -0.72, R) >droSim1.chrX_random 3861462 91 + 5698898 -----UUUUCUAGGGGGG--------UGGGGCUGGAGGUGCGCGU----UUUUCCAGAGCGUGCCAAGCUACGUGUGAAAAUAAUGAGCAGGCAUGCCAAACACAAAG- -----............(--------((...(((((((.......----.))))))).(((((((..(((...(((....)))...))).)))))))....)))....- ( -28.60, z-score = -0.67, R) >droVir3.scaffold_12970 11379920 88 + 11907090 ------------GCCAGA---UGGGGCGGGAGCAGGCGGGCCGGGAG-----CCUAGAGCGUGCCAAGCAACGUGUGAAAAUAAUGAGCGGGCAUGCUCAACACAAAG- ------------(((...---...)))(((..(.((....))..)..-----))).(((((((((..((....(((....)))....)).))))))))).........- ( -32.20, z-score = -1.38, R) >droGri2.scaffold_15203 3363890 94 + 11997470 ---------------GUACCAGGGUAAGCGGGUAGGUGUACCGAGAGGGGUAGCCGGAGCGUGCCAAGCAACGUGUGAAAAUAAUGAGCAGGCAUGCUUAACACAAUAG ---------------.((((.(......).))))(((...((....))....))).(((((((((..((....(((....)))....)).))))))))).......... ( -29.10, z-score = -1.71, R) >consensus _____UUUUCUAGGGGGG___GGGGGUGGGGGUGGAGGUGCGGGU____UUUUCCAGAGCGUGCCAAGCUACGUGUGAAAAUAAUGAGCAGGCAUGCCAAACACAAAG_ ..........................................................(((((((..((....(((....)))....)).)))))))............ (-13.35 = -14.73 + 1.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:04 2011