| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,561,773 – 14,561,880 |
| Length | 107 |
| Max. P | 0.574444 |

| Location | 14,561,773 – 14,561,880 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 63.56 |
| Shannon entropy | 0.67553 |
| G+C content | 0.42877 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -5.38 |
| Energy contribution | -5.61 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.574444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14561773 107 - 22422827 CACACUCACACAUCGCAUAAAGAGGACACUAACAAAGUCUUUUGUUUUGCUUUGAAGACUUUGCUG-------GUGCGAGUGUGUUAAUAUUGUAUGUAUACGUAUACGAACAC (((((((.....((.......)).(.(((((.((((((((((.(.....)...)))))))))).))-------)))))))))))......(((((((......))))))).... ( -31.40, z-score = -2.75, R) >droSim1.chrX_random 3853536 102 - 5698898 CACACUCACACAUCGCAUAAAGAGGACACUAACAAAGUCUUUUGUUUUGCUUUGAAGACUUUGCUG-------GUGCGAGUGUGUUAA-----UAUGUAUACGUAUAAGAACAC (((((((.....((.......)).(.(((((.((((((((((.(.....)...)))))))))).))-------)))))))))))...(-----((((....)))))........ ( -29.50, z-score = -2.73, R) >droEre2.scaffold_4690 6328058 105 + 18748788 ---ACCCACACAGCCCACAAAGAGGACACUAACAAAGUCCUUUGUUUUGCUUUGAAGACUUGGCUGCUGCAAAGUGUGAGUGUGUUAA-----UUUGUACACACGUAC-AAUAC ---.....(((((((.....(((((((.........)))))))(((((......)))))..))))).)).....((((.((((((...-----.....)))))).)))-).... ( -28.20, z-score = -1.06, R) >droAna3.scaffold_12905 505658 85 + 569613 CACCUGUACGCCCCCCACAAUGAGGACAGUAA-AAAGCUCUUUGUGCGGGCUUGGAG-CGUGGU---------GGUUCUGUGUGCUUACAGGACCU------------------ (((((.((.((((..(((((.(((........-....))).))))).)))).)).))-.)))..---------(((((((((....))))))))).------------------ ( -32.80, z-score = -1.95, R) >dp4.Unknown_group_97 31838 95 - 51274 ------GCCACCCCCCAUUAAAAGGACACUAACAAAGUCUUUUGUGCCGUUUUGAAGACUUUACUUGU-----AUUUUAAUACGUAUACGC--CCCGUAUACACACAC------ ------..........(((((((..(((.....(((((((((...........)))))))))...)))-----.)))))))..(((((((.--..)))))))......------ ( -18.00, z-score = -2.54, R) >droPer1.super_17 265765 101 + 1930428 GCCACCCCCACCCCCCAUUAAAAGGACACUAACAAAGUCUUUUGUGCCGUUUUGAAGACUUUACUUGU-----AUUUUAAUACGUAUACGC--CCCGUAUACACACAC------ ................(((((((..(((.....(((((((((...........)))))))))...)))-----.)))))))..(((((((.--..)))))))......------ ( -18.00, z-score = -2.83, R) >droSec1.super_305 9603 98 - 17228 CACACUCACACAUCGCAUAAAGAGGACACUAACAAAGUCUUUUGUUUUGCUUUGAAGACUUUGCUG-------GUGCGAGUGUGUCA------UAUGUGUACGUAUACGAA--- (((((((.....((.......)).(.(((((.((((((((((.(.....)...)))))))))).))-------)))))))))))..(------((((....))))).....--- ( -29.50, z-score = -2.12, R) >consensus CACACUCACACACCCCAUAAAGAGGACACUAACAAAGUCUUUUGUUUUGCUUUGAAGACUUUGCUG_______GUGUGAGUGUGUUAA_____UAUGUAUACACAUAC_AA___ ......(((((.........(((((((.........))))))).....((............))...............))))).............................. ( -5.38 = -5.61 + 0.23)

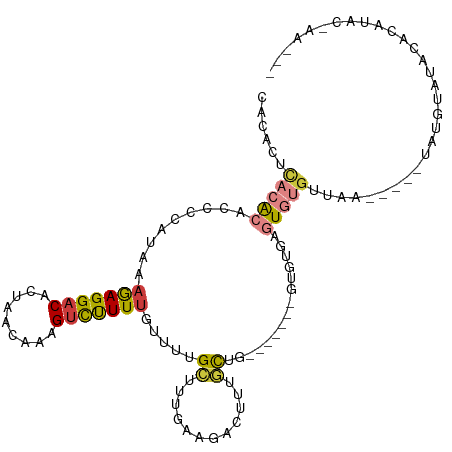

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:03 2011