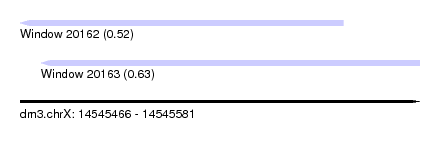
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,545,466 – 14,545,581 |
| Length | 115 |
| Max. P | 0.631697 |

| Location | 14,545,466 – 14,545,559 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Shannon entropy | 0.41786 |
| G+C content | 0.51434 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.55 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
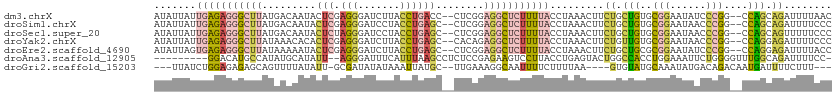
>dm3.chrX 14545466 93 - 22422827 CAAUACUCGAGGGAUCUUACCUGACC--CUCGGAGGCUCUUUUACCUAAACUUCUGCUGUGCGGAAUAUCCCGG--CCAGCAGAUUUUAACGGGACA .....((((((((.((......))))--))).))).........(((.....(((((((..(((......))).--.))))))).......)))... ( -31.50, z-score = -3.01, R) >droSim1.chrX 11218131 93 - 17042790 CAAUACUCGAGGGAUCCUACCUGAGC--CUCGGAGGCUCUUUUACCUAAACUUCUGCUGUGCGGAAUAACCCGG--CCAGCAGAUUUUCCCGGCACA ..........((((........((((--(.....))))).............(((((((..(((......))).--.)))))))...))))...... ( -30.90, z-score = -2.23, R) >droSec1.super_20 1130984 93 - 1148123 CAAUACUCUAGGGAUCCUACCUGAGC--CUCGGAGGCUCUUUUACCUAAACUUCUGCUGUGCGGAAUAACCCGG--CCAGCAGUUUUUCCCGGCACA ..........((((........((((--(.....)))))..............((((((..(((......))).--.))))))....))))...... ( -28.90, z-score = -1.69, R) >droYak2.chrX 8773681 93 - 21770863 ACACACUCGAGGGAUCUUACCUGAGC--CACAGAGGCUCUUUUACCUAAACUUCUGUUGUGCGGAAUAACCCGG--CCAGGAGAUUUUCCCGCGACA ......(((.((((..((.((((.((--((((((((..............))))))).....((......))))--))))).))...)))).))).. ( -29.34, z-score = -2.48, R) >droEre2.scaffold_4690 6313490 93 + 18748788 AAAUACUCGAGGGAUCUUACCUGAGC--CUCGGAGGCUCUUUUACCUAAACUUCUGCUGCGCGGAAUAUCCCGG--CCAGGAGAUUUUACCGGGACA .....((((.(((((((..((((.((--(..(((((........)).....((((((...))))))..))).))--))))))))))))..))))... ( -27.60, z-score = -0.74, R) >droAna3.scaffold_12905 474085 93 + 569613 ---CAUAUUAGGGAUUUCAUUUAAGCCUCUCCGAGAAGUCCUUACCUGAGUACUGGCCACCUGGAAAUUCUGGGGUUUGGCAGAUUUU-CCGGGCCA ---.....((((((((((................))))))))))..........((((....(((((.((((........)))).)))-)).)))). ( -27.89, z-score = -0.33, R) >consensus CAAUACUCGAGGGAUCUUACCUGAGC__CUCGGAGGCUCUUUUACCUAAACUUCUGCUGUGCGGAAUAACCCGG__CCAGCAGAUUUUCCCGGGACA ........((((((.(((..(((((...)))))))).)))))).((......(((((((..(((......)))....))))))).......)).... (-14.32 = -14.55 + 0.23)
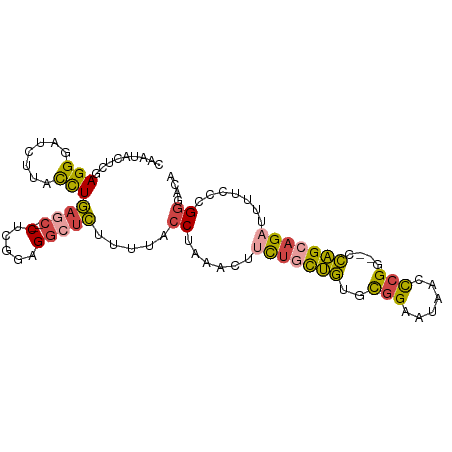
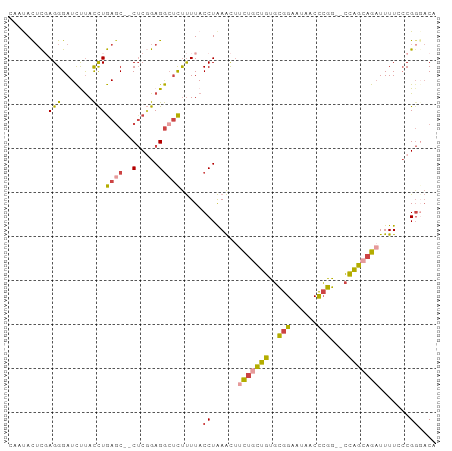
| Location | 14,545,472 – 14,545,581 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 65.70 |
| Shannon entropy | 0.68427 |
| G+C content | 0.44200 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -9.53 |
| Energy contribution | -10.74 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.631697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14545472 109 - 22422827 AUAUUAUUGAGAGGGCUUAUGACAAUACUCGAGGGAUCUUACCUGACC--CUCGGAGGCUCUUUUACCUAAACUUCUGCUGUGCGGAAUAUCCCGG--CCAGCAGAUUUUAAC .......(((((((((((..........(((((((.((......))))--))))))))))))))))..((((..(((((((..(((......))).--.))))))).)))).. ( -38.90, z-score = -4.25, R) >droSim1.chrX 11218137 109 - 17042790 AUAUUAUUGAGAGGGCUUAUGACAAUACUCGAGGGAUCCUACCUGAGC--CUCGGAGGCUCUUUUACCUAAACUUCUGCUGUGCGGAAUAACCCGG--CCAGCAGAUUUUCCC .......(((((((((((.(((.....(((.(((.......)))))).--.))).)))))))))))........(((((((..(((......))).--.)))))))....... ( -34.40, z-score = -2.07, R) >droSec1.super_20 1130990 109 - 1148123 AUAUUAUUGAGAGGGCUUAUGACAAUACUCUAGGGAUCCUACCUGAGC--CUCGGAGGCUCUUUUACCUAAACUUCUGCUGUGCGGAAUAACCCGG--CCAGCAGUUUUUCCC .......(((((((((((.(((.....(((.(((.......)))))).--.))).))))))))))).........((((((..(((......))).--.))))))........ ( -32.40, z-score = -1.41, R) >droYak2.chrX 8773687 109 - 21770863 AUAUUAUUGAGAGGGCUUAUAAACACACUCGAGGGAUCUUACCUGAGC--CACAGAGGCUCUUUUACCUAAACUUCUGUUGUGCGGAAUAACCCGG--CCAGGAGAUUUUCCC ......(((((.(............).)))))((((..((.((((.((--((((((((..............))))))).....((......))))--))))).))...)))) ( -27.64, z-score = -0.62, R) >droEre2.scaffold_4690 6313496 109 + 18748788 AUAUUAGUGAGAGGGCUUAUAAAAAUACUCGAGGGAUCUUACCUGAGC--CUCGGAGGCUCUUUUACCUAAACUUCUGCUGCGCGGAAUAUCCCGG--CCAGGAGAUUUUACC ...(((((((((((((((..........((((((..((......)).)--)))))))))))))))).)))).((((((..((.(((......))))--)))))))........ ( -35.10, z-score = -1.94, R) >droAna3.scaffold_12905 474091 101 + 569613 ---------GGACAUGCCAUAUGCAUAUU--AGGGAUUUCAUUUAAGCCUCUCCGAGAAGUCCUUACCUGAGUACUGGCCACCUGGAAAUUCUGGGGUUUGGCAGAUUUUCC- ---------((....(((((((.((...(--(((((((((................))))))))))..)).))).))))..)).(((((.((((........)))).)))))- ( -25.79, z-score = 0.27, R) >droGri2.scaffold_15203 3333244 100 - 11997470 ---UUAUCUGGAGAGAGCAGUUUUAUAUU-GCGAUAUAUAAAUUAUGC--UUGAAAGGCAAUUUUCUUUUAA----GUGUAUGCAAAUAUGACAGACAAUGAUUUUCUUU--- ---...((((......(((((.....)))-))..(((((....(((((--((((((((......))))))))----))))).....))))).))))..............--- ( -20.60, z-score = -1.16, R) >consensus AUAUUAUUGAGAGGGCUUAUAACAAUACUCGAGGGAUCUUACCUGAGC__CUCGGAGGCUCUUUUACCUAAACUUCUGCUGUGCGGAAUAACCCGG__CCAGCAGAUUUUCCC .......(((((((((((.........(((.(((.......))))))........))))))))))).........((.(((..(((......)))....))).))........ ( -9.53 = -10.74 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:03 2011