
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,519,854 – 14,519,944 |
| Length | 90 |
| Max. P | 0.989591 |

| Location | 14,519,854 – 14,519,944 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 80.41 |
| Shannon entropy | 0.37550 |
| G+C content | 0.38056 |
| Mean single sequence MFE | -19.19 |
| Consensus MFE | -15.01 |
| Energy contribution | -14.36 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
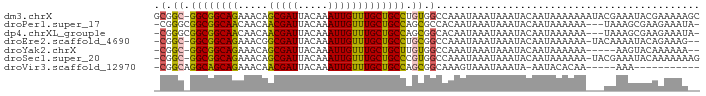
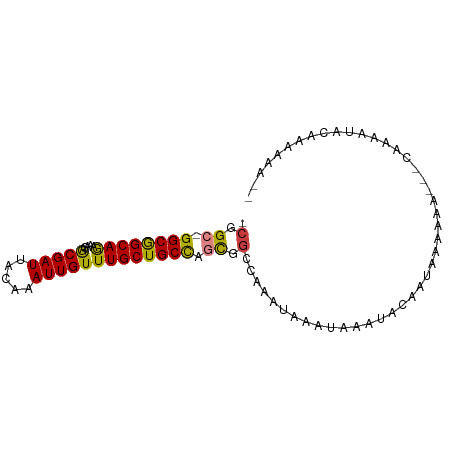
>dm3.chrX 14519854 90 + 22422827 GCGGC-GGCGGCAGAAACAGCGAUUACAAAUUGUUUGCUGCCUGUGGCCAAAUAAAUAAAUACAAUAAAAAAAUACGAAAUACGAAAAAGC (((((-.(((((((....((((((.....))))))..))))).)).)))..........................((.....)).....)) ( -20.90, z-score = -2.07, R) >droPer1.super_17 226582 86 - 1930428 -CGGGCGGCGGCAACAACAACGAUUACAAAUUGUUUGCUGCCAGCGCCACAAUAAAUAAAUACAAUAAAAAA---UAAAGCGAAGAAAUA- -.((((((((((((.....(((((.....))))))))))))).)).))........................---...............- ( -19.10, z-score = -2.25, R) >dp4.chrXL_group1e 7775136 86 - 12523060 -CGGGCGGCGGCAACAACAACGAUUACAAAUUGUUUGCUGCCAGCGGCACAAUAAAUAAAUACAAUAAAAAA---UAAAGCGAAGAAAUA- -...((.((((((.(((..(((((.....)))))))).)))).)).))........................---...............- ( -16.10, z-score = -1.23, R) >droEre2.scaffold_4690 6292820 86 - 18748788 -CGGC-GGCGGCAGAAACGGCGAUUACAAAUUGUUUGCUGCCUGCGGCCAAAUAAAUAAAUACAAUAAAAAA-UACAAAAUACAGAAAG-- -.(((-.(((((((....((((((.....))))))..))))).)).))).......................-................-- ( -21.90, z-score = -2.99, R) >droYak2.chrX 8752500 82 + 21770863 -CGGC-GGCGGCAGAAACAGCGAUUACAAAUUGUUUGCUGCUUGUGGCCAAAUAAAUAAAUACAAUAAAAAA-----AAGUACAAAAAA-- -.(((-.(((((((....((((((.....))))))..)))).))).))).......................-----............-- ( -18.50, z-score = -1.98, R) >droSec1.super_20 1110878 88 + 1148123 -CGGC-GGCGGCAGAAACAGCGAUUACAAAUUGUUUGCUGCCCGUGGCCAAAUAAAUAAAUACAAUAAAAAA-UACGAAAUACAAAAAAAG -.(((-.(((((((....((((((.....))))))..))).)))).))).......................-.................. ( -20.70, z-score = -2.85, R) >droVir3.scaffold_12970 11332403 73 + 11907090 -CGGCAGGCAGCAGAAACAACGAUUACAAAUUGUUUGCUGCCAGCGGCAAAGUAAAUAAAUA-AAUACACAA-----AAA----------- -(.((.(((((((.....((((((.....))))))))))))).)).)...............-.........-----...----------- ( -17.10, z-score = -2.63, R) >consensus _CGGC_GGCGGCAGAAACAGCGAUUACAAAUUGUUUGCUGCCAGCGGCCAAAUAAAUAAAUACAAUAAAAAA___CAAAAUACAAAAAA__ ...((.((((((((.....(((((.....))))))))))))).)).............................................. (-15.01 = -14.36 + -0.65)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:59 2011