| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,510,132 – 14,510,242 |
| Length | 110 |
| Max. P | 0.529270 |

| Location | 14,510,132 – 14,510,242 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Shannon entropy | 0.45196 |
| G+C content | 0.46028 |
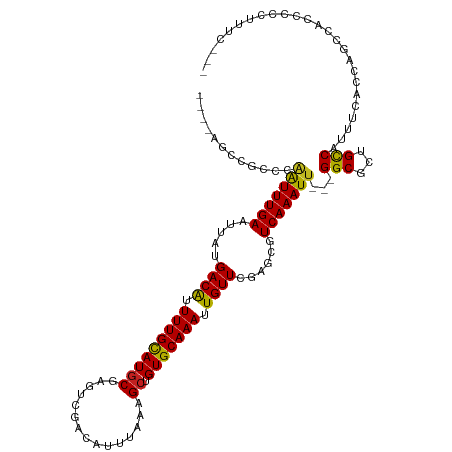
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.08 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
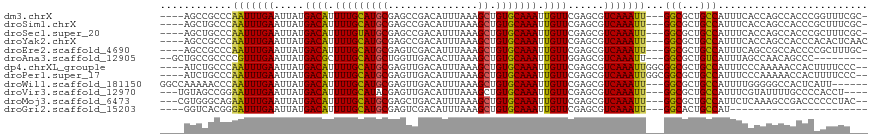
>dm3.chrX 14510132 110 - 22422827 ----AGCCGCCCAAUUUGAAUUAUGACAUUUUGCAUGCGAGCCGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU---GGCGCUGCCAUUUCACCAGCCACCCGGUUUCGC- ----.((.(((((((((((.....((((.(((((((((...............)).))))))).))))......)))))))---)).)).)).........((((....))))....- ( -30.86, z-score = -1.23, R) >droSim1.chrX 11178730 110 - 17042790 ----AGCUGCCCAAUUUGAAUUAUGACAUUUUGCAUGCGAGCCGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU---GGCGCUGCCAUUUCACCAGCCACCCGCUUUCGC- ----.((.(((((((((((.....((((.(((((((((...............)).))))))).))))......)))))))---)).)).)).........(((.....))).....- ( -29.36, z-score = -0.95, R) >droSec1.super_20 1101572 110 - 1148123 ----AGCUGCCCAAUUUGAAUUAUGACAUUUUGUAUGCGAGCCGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU---GGCGCUGCCAUUUCACCAGCCACCCGCUUUCGC- ----.((.(((((((((((.....((((.(((((((((...............)).))))))).))))......)))))))---)).)).)).........(((.....))).....- ( -27.36, z-score = -0.63, R) >droYak2.chrX 8743008 111 - 21770863 ----AGCCGCCCAAUUUGAAUUAUGACAUUUUGCAUGCGAGCCGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU---GGCGCUGCCAUUUCACCAGCCACCCACACUCAAC ----.((.(((((((((((.....((((.(((((((((...............)).))))))).))))......)))))))---)).)).)).......................... ( -28.66, z-score = -1.80, R) >droEre2.scaffold_4690 6283924 110 + 18748788 ----AGCCGCCCAAUUUGAAUUAUGACAUUUUGCAUGCGAGUCGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU---GGCGCUGCCAUUUCAGCCGCCACCCCGCUUUGC- ----.((.(((((((((((.....((((.(((((((((...............)).))))))).))))......)))))))---)).)).))..........((......)).....- ( -29.06, z-score = -0.71, R) >droAna3.scaffold_12905 444835 104 + 569613 --GCUGCCGCCCCGUUUGAAUUAUGACGCUUUGCAUGCUGGUUGACACUUAAAGCUGUGCAAAUUGUUGGAGCGUCAAAUU---GGCGCUGUCAUUUAGCCAACAGCCC--------- --((((..(((..((((((....(((((.((((((((((.............))).))))))).))))).....)))))).---)))((((.....))))...))))..--------- ( -30.52, z-score = -0.51, R) >dp4.chrXL_group1e 7764929 112 + 12523060 ----AUCUGCCCAAUUUGAAUUAUGACAUUUUGCAUGCGAGUUGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUUGGCGGCGCUGCCAUUUCCCAAAAACCACUUUUCCC-- ----....(((.(((((((.....((((.(((((((((...((((...)))).)).))))))).))))......))))))))))(((...))).......................-- ( -26.60, z-score = -1.02, R) >droPer1.super_17 216980 112 + 1930428 ----AUCUGCCCAAUUUGAAUUAUGACAUUUUGCAUGCGAGUUGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUUGGCGGCGCUGCCAUUUCCCAAAAACCACUUUUCCC-- ----....(((.(((((((.....((((.(((((((((...((((...)))).)).))))))).))))......))))))))))(((...))).......................-- ( -26.60, z-score = -1.02, R) >droWil1.scaffold_181150 642318 109 + 4952429 GGCCAAAAACCCAAUUUGAAUUAUGACAUUUUGCAUGCGAGUUGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU---GGCGCUGCCAUUUUGGGGGCCACUCAUU------ ((((.....(((((..((......((((.(((((((((...((((...)))).)).))))))).))))..((((((.....---))))))..))..))))))))).......------ ( -36.10, z-score = -2.41, R) >droVir3.scaffold_12970 11321883 108 - 11907090 ---UGUAGCCGGAAUUUGAAUUAUGACAUUUUGCAUACGAGUUGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU---GGCGCUGCCAUUUCGUAUUUUGCCCCACCU---- ---((..((.(((((.((((....((((.((((((((.(..((((...))))..))))))))).))))..((((((.....---)))))).....)))).)))))))..))...---- ( -26.10, z-score = -0.78, R) >droMoj3.scaffold_6473 6953107 110 + 16943266 ---CGUGGGCAGAAUUUGAAUUAUGACAUUUUGCAUGCGAGCUGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU---GGCGCUGCCAUUCUCAAAGCCGACCCCCCUAC-- ---...(((..(..(((((...((((((.(((((((...((((.........))))))))))).)))...((((((.....---))))))..)))..)))))..)..)))......-- ( -31.50, z-score = -1.47, R) >droGri2.scaffold_15203 3297540 88 - 11997470 ----GGUCACGGGAUUUGAAUUAUGACAUUUUGCAUGCGAGUCGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU---GGCACUGCCAU----------------------- ----........(((((((.....((((.(((((((((...............)).))))))).))))......)))))))---(((...)))..----------------------- ( -20.06, z-score = 0.48, R) >consensus ____AGCCGCCCAAUUUGAAUUAUGACAUUUUGCAUGCGAGUCGACAUUUAAAGCUGUGCAAAUUGUUCGAGCGUCAAAUU___GGCGCUGCCAUUUCACCAGCCACCCCCUUUC___ ............(((((((.....((((.(((((((((...............)).))))))).))))......)))))))...(((...)))......................... (-18.28 = -18.08 + -0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:56 2011