
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,489,980 – 14,490,096 |
| Length | 116 |
| Max. P | 0.611089 |

| Location | 14,489,980 – 14,490,096 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.82 |
| Shannon entropy | 0.54592 |
| G+C content | 0.46917 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -15.31 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14489980 116 - 22422827 CUCGCGUGUCAGUCGAGGGUUAAGCAAACACUUGUAGCCAAGUUCUUUGAUUGAUUCGUUGAGAAUGGAGGGGAUCGAAAUGGAAGCCGCAGAAGCCG-UGGUUGGUCAAUAACUAG ((((((.((((((((((((((.....)))(((((....))))).))))))))))).)).))))..(((....((((((.((((...(....)...)))-)..)))))).....))). ( -31.50, z-score = -0.48, R) >droSim1.chrX 11170471 107 - 17042790 CUCGCGUGUCAGUCGAGGGUUAAGCAAACACUUGUAACCAAGUUCUUUGAUUGAUUCGUUGCGAAUGGAGGGGCUCGGGAUG---------GAAGCCG-AGGUUGGUCAAUAGCUAG .(((((.((((((((((((((.....)))(((((....))))).)))))))))))....))))).(((..((.(((((....---------....)))-)).))..)))........ ( -32.30, z-score = -0.96, R) >droSec1.super_20 1081074 107 - 1148123 CUCGCGUGUCAGUCGAGGGUUAAGCAAACACUUGUAACCAAGUUCUUUGAUUGAUUCGUUGCGAAUGGAGGGGCUCGGAAUG---------GAAGCCG-AGGUUGGUCAAUAGUUAG .(((((.((((((((((((((.....)))(((((....))))).)))))))))))....))))).(((..((.(((((....---------....)))-)).))..)))........ ( -32.20, z-score = -1.52, R) >droYak2.chrX 8724049 112 - 21770863 CUCGCGUGUCAGUCGAGGGUUAAGCAAACACUUGUAACCAAGUUCUUUGAUUGACUCGUUGAGUAUGGAGGGGAUUGGAAUGGAAACGGGG-----AGCUGGUUGGACAAUAACUAG ((((((.((((((((((((((.....)))(((((....))))).))))))))))).)).))))......((..((((...(((...(((..-----..))).)))..))))..)).. ( -30.10, z-score = -0.99, R) >droEre2.scaffold_4690 6265053 116 + 18748788 CUCGCGUGUCAGUCGAGGGUUAAGCAAACACUUGUAACCAAGUUCCUUGAUUGAUUCGUUGAGUAUGGA-GGGAUCGGAAUGGAAACGGGGAAACCAGUUGGUUGGUCAAUAGCUAG ((((((.((((((((((((((.....)))(((((....))))).))))))))))).)).))))..(((.-..((((((..((....))((....))......)))))).....))). ( -36.30, z-score = -2.21, R) >droAna3.scaffold_13248 2222033 104 - 4840945 UUCGCGUGUCAG-AGACGACGAGGAAAGGGUUUUU-------UUUUUUUUUUGGGGCG--AAAGACACAAGUGUCAGAGGUA--AAUCGAUUCAUCAGUUAGUUACUGAAUCAAUU- (((((.((((..-.)))).(((((((((((.....-------)))))))))))..)))--)).((((....)))).......--....(((...(((((.....))))))))....- ( -22.90, z-score = -0.74, R) >consensus CUCGCGUGUCAGUCGAGGGUUAAGCAAACACUUGUAACCAAGUUCUUUGAUUGAUUCGUUGAGAAUGGAGGGGAUCGGAAUG__AAC_G__GAAGCAG_UGGUUGGUCAAUAACUAG ((((((.((((((((((((..........(((((....))))))))))))))))).)).)))).....................................(((((.....))))).. (-15.31 = -15.52 + 0.20)

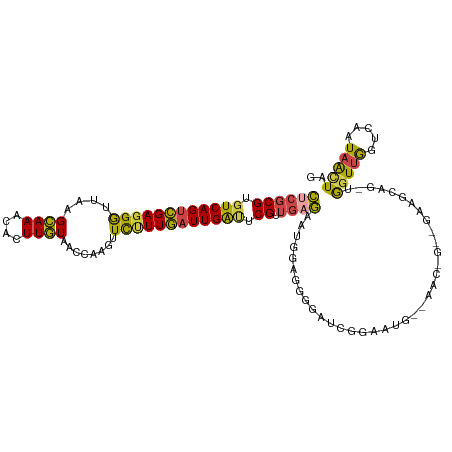
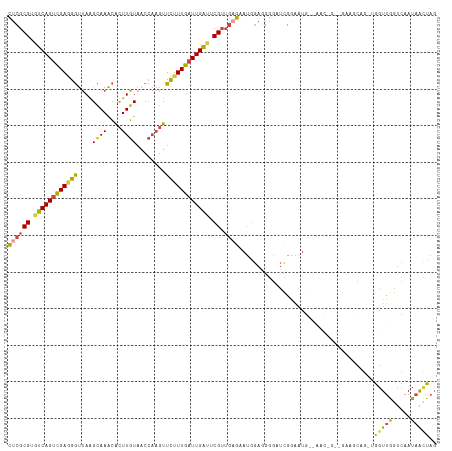
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:52 2011