| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,481,818 – 14,481,948 |
| Length | 130 |
| Max. P | 0.999322 |

| Location | 14,481,818 – 14,481,948 |
|---|---|
| Length | 130 |
| Sequences | 5 |
| Columns | 142 |
| Reading direction | forward |
| Mean pairwise identity | 63.74 |
| Shannon entropy | 0.61938 |
| G+C content | 0.38118 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
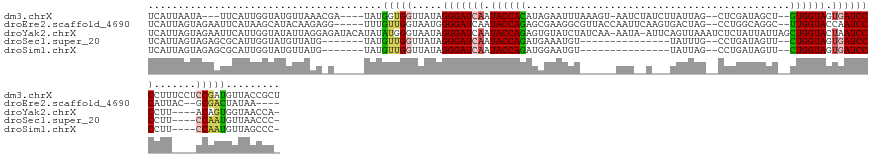
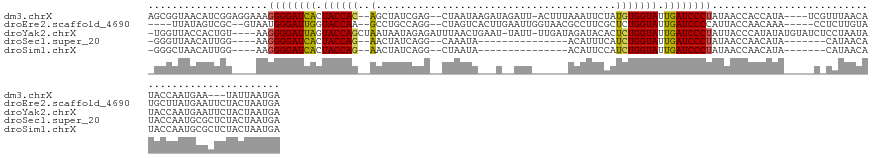
>dm3.chrX 14481818 130 + 22422827 UCAUUAAUA---UUCAUUGGUAUGUUAAACGA----UAUGGUGGUUAUAGGGAUCAAUACCACAUAGAAUUUAAAGU-AAUCUAUCUUAUUAG--CUCGAUAGCU--GUGGUAGUGAUCCCCUUUCCUCCGAUGUUACCGCU ...((((((---(........)))))))..((----(((((.((.....(((((((.(((((((((((.........-..)))))......((--((....))))--)))))).)))))))....)).)).)))))...... ( -34.70, z-score = -2.36, R) >droEre2.scaffold_4690 6257592 127 - 18748788 UCAUUAGUAGAAUUCAUAAGCAUACAAGAGG-----UUUGUUGGUAAUGGGGAUCAAUACCAGAGCGAAGGCGUUACCAAUUCAAGUGACUAG--CCUGGCAGGC--UUGGUACCAAUCCCAUUAC--GCGACUAUAA---- ......((((........((((.((.....)-----).)))).(((((((((.....((((((.((....))(((((........)))))..(--((.....)))--))))))....)))))))))--....))))..---- ( -34.10, z-score = -1.30, R) >droYak2.chrX 8716394 135 + 21770863 UCAUUAGUAGAAUUCAUUGGUAUAUUAGGAGAUACAUAUAUGGGUAAUAGGGAUCAAUACCAGAGUGUAUCUAUCAA-AAUA-AUUCAGUUAAAUCUCUAUUAUUAGCUGGUACUAAUCCCCUU----ACAGUGGUAACCA- (((((.((((.....((((((((....((((((((((...(((................)))..)))))))).))..-....-...(((((((..........)))))))))))))))....))----)))))))......- ( -26.19, z-score = 0.24, R) >droSec1.super_20 1073251 111 + 1148123 UCAUUAGUAGAGCGCAUUGGUAUGUUAUG-------UAUGUUGGUUAUAGGGAUCAAUACCAGAUGAAAUGU---------------UAUUUG--CCUGAUAGUU--CUGGUAGUGAUCCCCUU----CCAAUGUUAACCC- .............(((((((.......((-------((.......))))(((((((.(((((((.....(((---------------((....--..)))))..)--)))))).)))))))...----)))))))......- ( -31.10, z-score = -2.38, R) >droSim1.chrX 11162646 111 + 17042790 UCAUUAGUAGAGCGCAUUGGUAUGUUAUG-------UAUGUUGGUUAUAGGGAUCAAUACCAGAUGGAAUGU---------------UAUUAG--CCUGAUAGUU--CUGGUAGUGAUCCCCUU----CCAAUGUUAGCCC- ...........(((((((((.......((-------((.......))))(((((((.(((((((.....(((---------------((....--..)))))..)--)))))).)))))))...----)))))))..))..- ( -32.30, z-score = -2.13, R) >consensus UCAUUAGUAGAAUUCAUUGGUAUGUUAGG_G_____UAUGUUGGUUAUAGGGAUCAAUACCAGAUGGAAUGU______AAU__A___UAUUAG__CCUGAUAGUU__CUGGUAGUGAUCCCCUU____CCAAUGUUAACCC_ .......................................(((((.....(((((((.((((((............................................)))))).))))))).......)))))......... (-16.22 = -16.58 + 0.36)
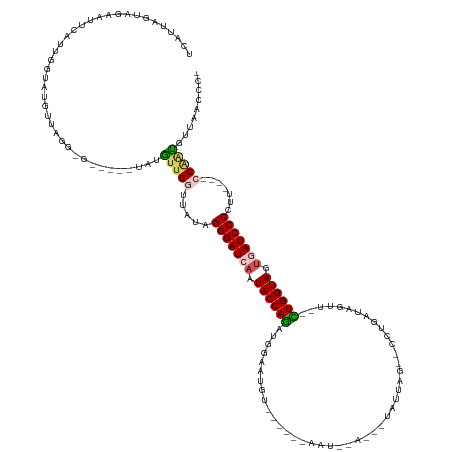
| Location | 14,481,818 – 14,481,948 |
|---|---|
| Length | 130 |
| Sequences | 5 |
| Columns | 142 |
| Reading direction | reverse |
| Mean pairwise identity | 63.74 |
| Shannon entropy | 0.61938 |
| G+C content | 0.38118 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.34 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
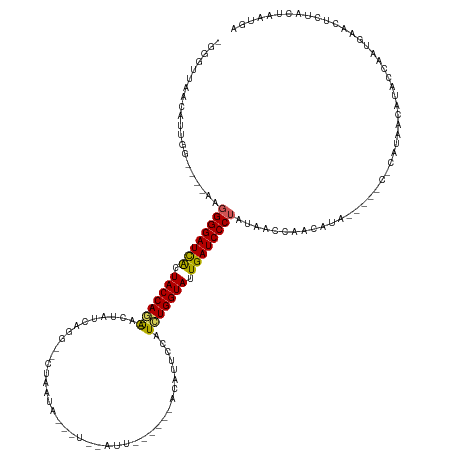
>dm3.chrX 14481818 130 - 22422827 AGCGGUAACAUCGGAGGAAAGGGGAUCACUACCAC--AGCUAUCGAG--CUAAUAAGAUAGAUU-ACUUUAAAUUCUAUGUGGUAUUGAUCCCUAUAACCACCAUA----UCGUUUAACAUACCAAUGAA---UAUUAAUGA ...((((.....((.((...((((((((.((((((--((((....))--))......(((((..-.........))))))))))).))))))))....)).))...----..(.....).))))......---......... ( -36.20, z-score = -3.60, R) >droEre2.scaffold_4690 6257592 127 + 18748788 ----UUAUAGUCGC--GUAAUGGGAUUGGUACCAA--GCCUGCCAGG--CUAGUCACUUGAAUUGGUAACGCCUUCGCUCUGGUAUUGAUCCCCAUUACCAACAAA-----CCUCUUGUAUGCUUAUGAAUUCUACUAAUGA ----((((((..((--((((((((((..(((((((--((.....(((--(..((.(((......))).))))))..))).))))))..)).))))))))..((((.-----....))))..))))))))............. ( -36.00, z-score = -2.40, R) >droYak2.chrX 8716394 135 - 21770863 -UGGUUACCACUGU----AAGGGGAUUAGUACCAGCUAAUAAUAGAGAUUUAACUGAAU-UAUU-UUGAUAGAUACACUCUGGUAUUGAUCCCUAUUACCCAUAUAUGUAUCUCCUAAUAUACCAAUGAAUUCUACUAAUGA -(((........((----((((((((((((((((((((....))).(((((....))))-)...-..............))))))))))))))).)))).....(((((........))))))))................. ( -27.00, z-score = -0.62, R) >droSec1.super_20 1073251 111 - 1148123 -GGGUUAACAUUGG----AAGGGGAUCACUACCAG--AACUAUCAGG--CAAAUA---------------ACAUUUCAUCUGGUAUUGAUCCCUAUAACCAACAUA-------CAUAACAUACCAAUGCGCUCUACUAAUGA -((((...((((((----..((((((((.((((((--(.........--......---------------........))))))).)))))))).((.......))-------.........)))))).))))......... ( -28.20, z-score = -3.21, R) >droSim1.chrX 11162646 111 - 17042790 -GGGCUAACAUUGG----AAGGGGAUCACUACCAG--AACUAUCAGG--CUAAUA---------------ACAUUCCAUCUGGUAUUGAUCCCUAUAACCAACAUA-------CAUAACAUACCAAUGCGCUCUACUAAUGA -((((...((((((----..((((((((.((((((--(.........--......---------------........))))))).)))))))).((.......))-------.........)))))).))))......... ( -29.80, z-score = -3.54, R) >consensus _GGGUUAACAUUGG____AAGGGGAUCACUACCAG__AACUAUCAGG__CUAAUA___U__AUU______ACAUUCCAUCUGGUAUUGAUCCCUAUAACCAACAUA_____C_CAUAACAUACCAAUGAACUCUACUAAUGA ....................((((((((.((((((............................................)))))).))))))))................................................ (-16.58 = -16.34 + -0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:50 2011