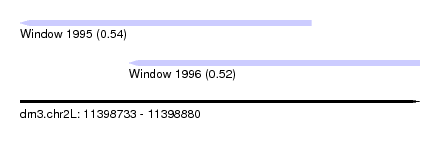
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,398,733 – 11,398,880 |
| Length | 147 |
| Max. P | 0.543310 |

| Location | 11,398,733 – 11,398,840 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 126 |
| Reading direction | reverse |
| Mean pairwise identity | 56.61 |
| Shannon entropy | 0.78245 |
| G+C content | 0.45345 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -5.57 |
| Energy contribution | -4.83 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
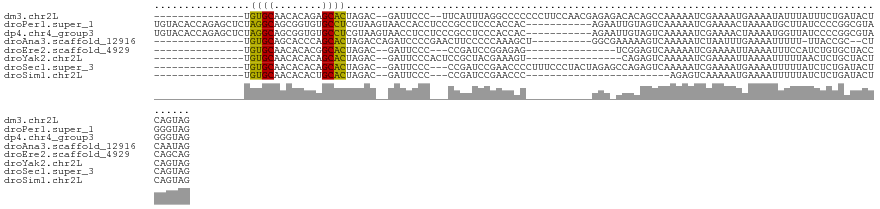
>dm3.chr2L 11398733 107 - 23011544 ---------------UGUGCAACACAGAGCACUAGAC--GAUUCCC--UUCAUUUAGGCCCCCCCUUCCAACGAGAGACACAGCCAAAAAUCGAAAAUGAAAAUAUUUAUUUCUGAUACUCAGUAG ---------------((((.........((.(((((.--((.....--.)).)))))))............(....).)))).......((((.(((((((....))))))).))))......... ( -13.90, z-score = -0.36, R) >droPer1.super_1 8297032 115 - 10282868 UGUACACCAGAGCUCUAGGCAGCGGUGUGCCUCGUAAGUAACCACCUCCCGCCUCCCACCAC-----------AGAAUUGUAGUCAAAAAUCGAAAACUAAAAUGCUUAUCCCCGGCGUAGGGUAG .(((((((...((.....))...))))))).............((((..((((........(-----------(.....((..((.......))..)).....)).........))))..)))).. ( -24.33, z-score = 0.19, R) >dp4.chr4_group3 6849139 115 - 11692001 UGUACACCAGAGCUCUAGGCAGCGGUGUGCCUCGUAAGUAACCUCCUCCCGCCUCCCACCAC-----------AGAAUUGUAGUCAAAAAUCGAAAACUAAAAUGGUUAUCCCCGGCGUAGGGUAG .(((((((...((.....))...)))))))............(((((..((((....((.((-----------......)).))........((.((((.....)))).))...))))..))).)) ( -25.20, z-score = 0.23, R) >droAna3.scaffold_12916 12107411 98 + 16180835 ---------------UGUGCAGCACCCAGCACUAGACCAGAUCCCCGAACUUCCCCCAAAGCU----------GGCGAAAAAGUCAAAAAUCUAAUUUGAAAAUUUUU-UUACCGC--CUCAAUAG ---------------.((((........))))...............................----------(((((((((.(((((.......)))))...)))))-....)))--)....... ( -13.40, z-score = -0.77, R) >droEre2.scaffold_4929 12600231 91 + 26641161 ---------------UGUGCAACACACGGCACUAGAC--GAUUCCC---CCGAUCCGGAGAG---------------UCGGAGUCAAAAAUCGAAAAUUAAAAUUUCCAUCUGUGCUACCCAGCAG ---------------.(((.....)))(((((.(((.--(((((.(---(......)).)))---------------))((((.....................)))).))))))))......... ( -19.70, z-score = -1.17, R) >droYak2.chr2L 7796913 93 - 22324452 ---------------UGUGCAACACACAGCACUAGAC--GAUUCCCACUCCGCUACGAAAGU----------------CAGAGUCAAAAAUCGAAAAUUAAAAUUUUUAACUCUGCUACUCAGUAG ---------------.((((........)))).....--.............((((...(((----------------((((((.((((((...........)))))).))))))..)))..)))) ( -17.20, z-score = -2.08, R) >droSec1.super_3 6792942 106 - 7220098 ---------------UGUGCAACACACAGCACUAGAC--GAUUCCC---CCGAUCCGAACCCCUUUCCCUACUAGAGCCAGAGUCAAAAAUCGAAAAUGAAAAUUUUUAUCUCUGAUACUCAGUAG ---------------.((((........))))....(--(..((..---..))..))...........(((((.(((.(((((..((((((...........))))))..)))))...)))))))) ( -19.80, z-score = -2.90, R) >droSim1.chr2L 11207156 82 - 22036055 ---------------UGUGCAACACACUGCACUAGAC--GAUUCCC---CCGAUCCGAACCC------------------------AGAGUCAAAAAUGAAAAUUUUUAUCUCUGAUACUCAGUAG ---------------.(((((......)))))....(--(..((..---..))..))....(------------------------((((..((((((....))))))..)))))........... ( -18.10, z-score = -3.59, R) >consensus _______________UGUGCAACACACAGCACUAGAC__GAUUCCC___CCGCUCCGAACAC___________AGA_UCAGAGUCAAAAAUCGAAAAUGAAAAUUUUUAUCUCUGAUACUCAGUAG ................((((........)))).............................................................................................. ( -5.57 = -4.83 + -0.75)

| Location | 11,398,773 – 11,398,880 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 60.15 |
| Shannon entropy | 0.77498 |
| G+C content | 0.58372 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -12.66 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.27 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
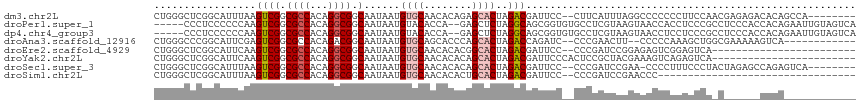
>dm3.chr2L 11398773 107 - 23011544 CUGGGCUCGGCAUUUAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACAGAGCACUAGACGAUUCC--CUUCAUUUAGGCCCCCCCUUCCAACGAGAGACACAGCCA-------- .((((((((((......))))).))).))((((((.......(((........)))(((((.((....--..)).))))))))...........(....).....))).-------- ( -27.70, z-score = -0.06, R) >droPer1.super_1 8297072 110 - 10282868 -----CCCUCCCCCCAAGUCGGCGCCACAGGCGGCAAUAAUGUACACCA--GAGCUCUAGGCAGCGGUGUGCCUCGUAAGUAACCACCUCCCGCCUCCCACCACAGAAUUGUAGUCA -----............((.((.(....((((((.......(((((((.--..((.....))...)))))))..(....)..........))))))..).))))............. ( -24.00, z-score = 0.99, R) >dp4.chr4_group3 6849179 110 - 11692001 -----CCCUCCCCCCAAGUCGGCGCCACAGGCGGCAAUAAUGUACACCA--GAGCUCUAGGCAGCGGUGUGCCUCGUAAGUAACCUCCUCCCGCCUCCCACCACAGAAUUGUAGUCA -----............((.((.(....((((((.......(((((((.--..((.....))...)))))))..(....)..........))))))..).))))............. ( -24.00, z-score = 0.96, R) >droAna3.scaffold_12916 12107448 101 + 16180835 CUGGGCCCGGCAUUCGAGUCGGCGCCACAGACGGCAAUAAUGUGCAGCACCCAGCACUAGACCAGAUC--CCCGAACUU--CCCCCAAAGCUGGCGAAAAAGUCA------------ (((((((((((......))))).))).)))..(((......((((........))))..(.((((...--.........--.........)))))......))).------------ ( -27.90, z-score = -0.09, R) >droEre2.scaffold_4929 12600271 91 + 26641161 CUGGGCUCGGCAUUCAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACGGCACUAGACGAUUCC--CCCGAUCCGGAGAGUCGGAGUCA------------------------ (((((((((((......))))).))).)))..(((......((((........))))....((((((.--((......)).))))))..))).------------------------ ( -31.50, z-score = -0.90, R) >droYak2.chr2L 7796953 93 - 22324452 CUGGGCUCGGCAUUCAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACAGCACUAGACGAUUCCCACUCCGCUACGAAAGUCAGAGUCA------------------------ ...((((((((......((((.((((...)))).)......((((........))))..)))(.....).....)))((....))..))))).------------------------ ( -26.50, z-score = -0.82, R) >droSec1.super_3 6792982 106 - 7220098 CUGGGCUCGGCAUUUAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACAGCACUAGACGAUUCC--CCCGAUCCGAA-CCCCUUUCCCUACUAGAGCCAGAGUCA-------- ...((((((((.((((.((((.((((...)))).)......((((........))))..)))((((..--...))))....-.............))))))).))))).-------- ( -29.30, z-score = -0.82, R) >droSim1.chr2L 11207196 82 - 22036055 CUGGGCUCGGCAUUUAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACUGCACUAGACGAUUCC--CCCGAUCCGAACCC--------------------------------- .((((.((((.......((((.((((...)))).)......(((((......)))))..)))......--.)))))))).....--------------------------------- ( -26.04, z-score = -1.41, R) >consensus CUGGGCUCGGCAUUCAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACAGCACUAGACGAUUCC__CCCGAUCAAGAAACCCCCUCCCA_C______CACA____________ .................((((.((((...)))).)......((((........))))..)))....................................................... (-12.66 = -13.10 + 0.44)
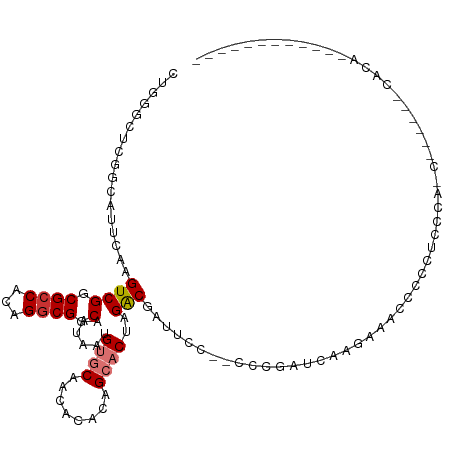
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:25 2011