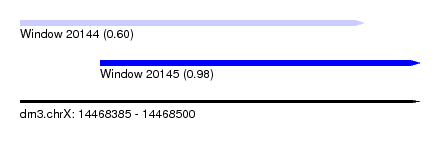
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,468,385 – 14,468,500 |
| Length | 115 |
| Max. P | 0.975110 |

| Location | 14,468,385 – 14,468,484 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Shannon entropy | 0.43428 |
| G+C content | 0.48771 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
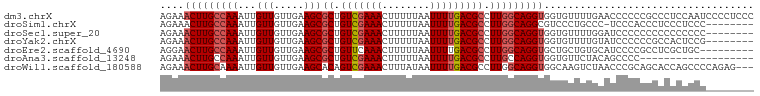
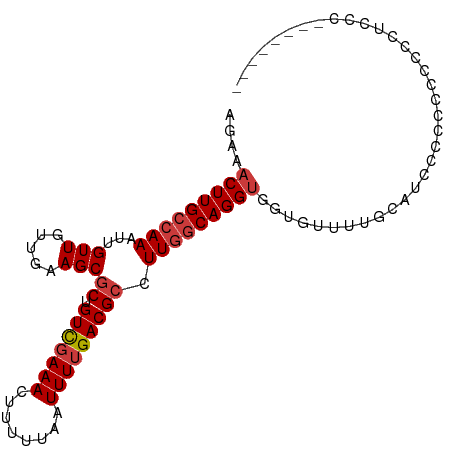
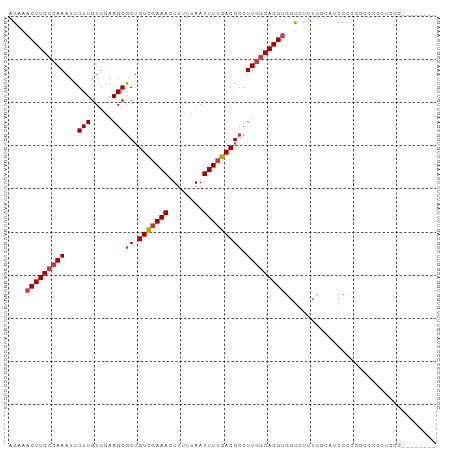
>dm3.chrX 14468385 99 + 22422827 AGAAACUUGCCAAAUUGUUGUUGAAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGUGGUGUUUUGAACCCCCCGCCCUCCAAUCCCCUCCC ....(((((((((...(((.....)))((.(((((((........))))))))).)))))))))((((............))))............... ( -22.60, z-score = -1.54, R) >droSim1.chrX 11148683 90 + 17042790 AGAAACUUGCCAAAUUGUUGUUGAAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGCGUCCCUGCCC-UCCCACCCUCCCUCCC-------- .((..((((((((...(((.....)))((.(((((((........))))))))).))))))))..)).......-................-------- ( -20.40, z-score = -1.92, R) >droSec1.super_20 1060096 91 + 1148123 AGAAACUUGCCAAAUUGUUGUUGAAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGUGGUGUUUUGGAUCCCCCCCCCCCCCCC-------- ....(((((((((...(((.....)))((.(((((((........))))))))).)))))))))((......((......))......)).-------- ( -21.10, z-score = -1.22, R) >droYak2.chrX 8702958 91 + 21770863 AGAAACUUGCCAAAUUGUUGUUGAAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGUGGUGUUUUGUAUCCCCCCGCCACUCCG-------- ....(((((((((...(((.....)))((.(((((((........))))))))).)))))))))((((.............))))......-------- ( -23.42, z-score = -1.75, R) >droEre2.scaffold_4690 6244905 90 - 18748788 AGGAACUUGCCAAAUUGUUGUUGAAGCGCUGUUCAAACUUUUUAAUUUUGACGCCUUGGCAGGUGCUGCUGUGCAUCCCCGCCUCGCUGC--------- .((((((((((((..(((((((((((.(.........).)))))))...))))..)))))))))((......)).)))............--------- ( -19.70, z-score = 1.27, R) >droAna3.scaffold_13248 2188984 80 + 4840945 AGAAACUUGCCAAAUUGUUGUUGAAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGCCAGGUGGUGUUCUACAGCCCC------------------- ................(((((.(((.(((((((((((........)))))))((((....))))))))))).)))))...------------------- ( -18.80, z-score = -0.94, R) >droWil1.scaffold_180588 251918 96 + 1294757 AGAAACUUGCAAAAUUGUUGUUGAAGCACAGUCGAAACUUUAUAAUUUUGACGCCUUGGCAGGUGGCAAGUCUAACCCGCAGCACCAGCCCCAGAG--- ....((((((.....((((.....))))..(((((((........)))))))......))))))(((..((((.......)).))..)))......--- ( -18.30, z-score = 1.11, R) >consensus AGAAACUUGCCAAAUUGUUGUUGAAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGUGGUGUUUUGCAUCCCCCCCCCCCUCCC________ ....(((((((((...(((.....)))((.(((((((........))))))))).)))))))))................................... (-16.26 = -16.86 + 0.59)
| Location | 14,468,408 – 14,468,500 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 67.96 |
| Shannon entropy | 0.54313 |
| G+C content | 0.55507 |
| Mean single sequence MFE | -18.74 |
| Consensus MFE | -11.26 |
| Energy contribution | -11.14 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14468408 92 + 22422827 AAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGUGGUGUUUUGAACCCCCCGCCCUCCAAUC----CCCUCCCCUCGUCGACUCCCACC (((.((.(((((((........))))))))))))(((.((.(((.......))).)).)))........----....................... ( -19.20, z-score = -1.15, R) >droSim1.chrX 11148706 77 + 17042790 AAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGCGUC--CCUGCCCUCCCACCCUCCCUCCC----CUCUCCUACC------------- (((.((.(((((((........))))))))))))((((((....--)))))).................----..........------------- ( -18.40, z-score = -3.83, R) >droSec1.super_20 1060119 78 + 1148123 AAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGUGGUGUUUUGGAUCCCCC-CCCCCCCCCC----CUCUCUUACC------------- (((.((.(((((((........))))))))))))((.(((.((......((......)-)......)))----)).)).....------------- ( -14.20, z-score = 0.05, R) >droYak2.chrX 8702981 81 + 21770863 AAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGUGGUGUUUUGUAUCCCCC-CGCCACUCCG----CCCACCUACCACC---------- (((.((.(((((((........))))))))))))(((.(((((((.............-)))))))..)----))...........---------- ( -21.12, z-score = -2.16, R) >droWil1.scaffold_180588 251941 96 + 1294757 AAGCACAGUCGAAACUUUAUAAUUUUGACGCCUUGGCAGGUGGCAAGUCUAACCCGCAGCACCAGCCCCAGAGCAACACCAUCAGCAACACCCACC ..((...(((((((........)))))))((((.(((.((((((...........))..)))).)))..)).))..........)).......... ( -20.80, z-score = -1.11, R) >consensus AAGCGCUGUCGAAACUUUUUAAUUUUGACGCCUUGGCAGGUGGUGUUUUGAACCCCCC_CCCCCCCCCC____CCCUCCUACC__C__________ ...(((((((((((........)))))))((....)).))))...................................................... (-11.26 = -11.14 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:48 2011