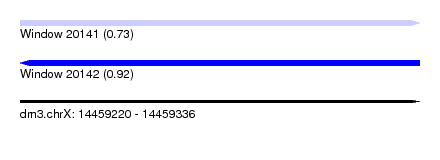
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,459,220 – 14,459,336 |
| Length | 116 |
| Max. P | 0.916197 |

| Location | 14,459,220 – 14,459,336 |
|---|---|
| Length | 116 |
| Sequences | 9 |
| Columns | 138 |
| Reading direction | forward |
| Mean pairwise identity | 75.13 |
| Shannon entropy | 0.47567 |
| G+C content | 0.44184 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.01 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
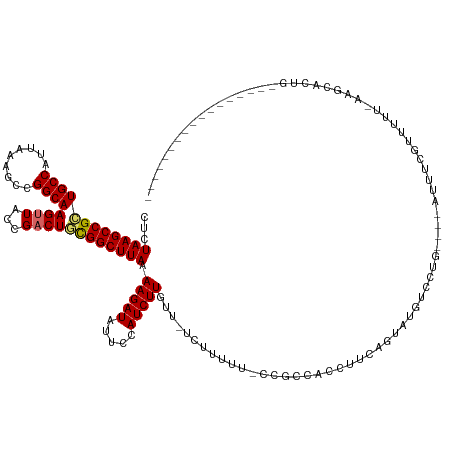
>dm3.chrX 14459220 116 + 22422827 CUCUAAGCCGCUGCCAUUAAAGCCGGCAAGUUACCGACUGCGGCUUAAAAGAUAUUCCAUCUUGUU-UCUUUUUUCCGCCACCUUCAGAAUGUCCUG----AUUUCGUUUUUUCAGCACUG----------------- ...((((((((((((.........))))((((...))))))))))))...(((((((.........-....................)))))))(((----(..........)))).....----------------- ( -23.45, z-score = -1.10, R) >droSim1.chrX 11140541 114 + 17042790 CUCUAAGCCGCUGCCAUUAAAGCCGGCAAGUUACCGACUGCGGCUUAAAAGAUAUUCCAUCUUGUU-UCUUUUU-CCGCCACCUUCAGUUUGUCCUG----AUUUCGUUUUU-CAGCACUG----------------- ...((((((((((((.........))))((((...)))))))))))).(((((.....)))))...-.......-................((.(((----(.........)-))).))..----------------- ( -23.30, z-score = -1.16, R) >droSec1.super_20 1049369 114 + 1148123 CUCUAAGCCGCUGCCAUUAAAGCCGGCAAGUUACCGACUGCGGCUUAAAAGAUAUUCCAUCUUGUU-UCUUUUU-CCGCCACCUUCAGAAUGUCCUG----AUUUCGUUUUU-CAGCACUG----------------- ...((((((((((((.........))))((((...))))))))))))...(((((((.........-.......-............)))))))(((----(.........)-))).....----------------- ( -23.59, z-score = -1.12, R) >droYak2.chrX 8693565 113 + 21770863 CUCUAAGCCGCUGCCAUUAAAGCCGGCAAGUUACCGACUGCGGCUUAAAAGAUAUUCCAUCUUGUU-UCUUUUU-CCGCCACCUUCAGAAUGUCCUG----AUUUCGUUUUU--AGCACUG----------------- ......((((((........)).))))..((((..(((.((((...((((((.((........)).-)))))).-)))).....((((......)))----)....)))..)--)))....----------------- ( -25.20, z-score = -1.72, R) >droEre2.scaffold_4690 6237182 112 - 18748788 CUCUAAGCCGCUGCCAUUAAAGCCGGCAAGUUACCGACUGCGGCUUAAAAGAUAUUCCAUCUUGUU-UCUUUGC--CGCCACCUUCAGAAUGUCCUG----AUUUCGUUUUU--AGCGCUG----------------- ......((((((........)).))))..((((..(((.(((((...(((((.((........)).-)))))))--))).....((((......)))----)....)))..)--)))....----------------- ( -26.70, z-score = -1.26, R) >droAna3.scaffold_13248 2179587 132 + 4840945 CUCUAAGCCGUUGCCAUUAAAGCCGGCAAGUUACCGACUACGGCUUAAAAGAUAUUCCAUCUUGUU-UCUUUUG-CCGCCACCUUCGGUUCGUUCUG----AUUUUCUUUUUUAAAUGGUGGCACAUGGGUGGCAUGA ......((((..((.......))))))..((((((......(((..((((((.((........)).-)))))))-))((((((.((((......)))----)...............)))))).....)))))).... ( -35.89, z-score = -1.26, R) >dp4.chrXL_group1e 7706159 131 - 12523060 CUCUAAGCCGCUGCCAUUAAAGCCGGCAAGCUACCGACUAUGGCUUAAAAGAUAUUCCAUCUUGUUCUCAUCUUUGGGGCACCCUUCCUCCCACAGAGGGUAUGACUGUUUUCAGCUAAUUUUUGAUAUAA------- ......(((((((((.........))).)))..((((..((((.....(((((.....)))))....))))..)))))))((((((.........))))))....(((....)))................------- ( -28.60, z-score = 0.37, R) >droPer1.super_17 157346 131 - 1930428 CUCUAAGCCGCUGCCAUUAAAGCCGGCAAGCUACCGACUAUGGCUUAAAAGAUAUUCCAUCUUGUUCUCAUCUUUGGGGCACCCUUCCUCCUACAGAGGGUAUGACUAUUUUCAGCUAACUUUUGAUAUAA------- ......(((((((((.........))).)))..((((..((((.....(((((.....)))))....))))..)))))))((((((.........))))))..............................------- ( -27.60, z-score = 0.32, R) >droWil1.scaffold_180588 243322 120 + 1294757 CUCUAAGCCGUUGCCAUUAAAGCCGGCAAGUUACCGACUACGGCUUAAAAGAUAUUCCAUCUUGUC-UUAUUUU------CUUUUUCCUUUUUUUUG----UCCUCCUUCUUGUGACAUUUUCUAAUAUGU------- ...((((((((((((.........))))((((...)))))))))))).((((((........))))-)).....------...............((----(((........).)))).............------- ( -22.20, z-score = -2.70, R) >consensus CUCUAAGCCGCUGCCAUUAAAGCCGGCAAGUUACCGACUGCGGCUUAAAAGAUAUUCCAUCUUGUU_UCUUUUU_CCGCCACCUUCAGUAUGUCCUG____AUUUCGUUUUU_AAGCACUG_________________ ...((((((((((((.........))))((((...)))))))))))).(((((.....)))))........................................................................... (-16.99 = -17.01 + 0.03)


| Location | 14,459,220 – 14,459,336 |
|---|---|
| Length | 116 |
| Sequences | 9 |
| Columns | 138 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Shannon entropy | 0.47567 |
| G+C content | 0.44184 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -22.01 |
| Energy contribution | -21.42 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
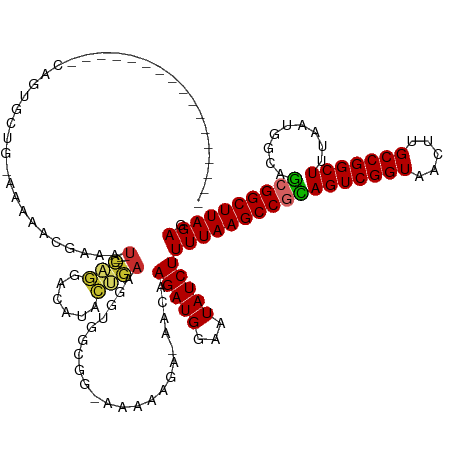
>dm3.chrX 14459220 116 - 22422827 -----------------CAGUGCUGAAAAAACGAAAU----CAGGACAUUCUGAAGGUGGCGGAAAAAAGA-AACAAGAUGGAAUAUCUUUUAAGCCGCAGUCGGUAACUUGCCGGCUUUAAUGGCAGCGGCUUAGAG -----------------..((.((((..........)----))).))(((((....((..(........).-.)).....)))))....((((((((((...((((.....))))(((.....))).)))))))))). ( -32.50, z-score = -1.81, R) >droSim1.chrX 11140541 114 - 17042790 -----------------CAGUGCUG-AAAAACGAAAU----CAGGACAAACUGAAGGUGGCGG-AAAAAGA-AACAAGAUGGAAUAUCUUUUAAGCCGCAGUCGGUAACUUGCCGGCUUUAAUGGCAGCGGCUUAGAG -----------------..((.(((-(.........)----))).))..(((((..(((((..-.((((((-..............))))))..)))))..)))))..((.(((((((.....)))..))))..)).. ( -32.44, z-score = -2.00, R) >droSec1.super_20 1049369 114 - 1148123 -----------------CAGUGCUG-AAAAACGAAAU----CAGGACAUUCUGAAGGUGGCGG-AAAAAGA-AACAAGAUGGAAUAUCUUUUAAGCCGCAGUCGGUAACUUGCCGGCUUUAAUGGCAGCGGCUUAGAG -----------------..((.(((-(.........)----))).))(((((....((..(..-.....).-.)).....)))))....((((((((((...((((.....))))(((.....))).)))))))))). ( -32.30, z-score = -1.65, R) >droYak2.chrX 8693565 113 - 21770863 -----------------CAGUGCU--AAAAACGAAAU----CAGGACAUUCUGAAGGUGGCGG-AAAAAGA-AACAAGAUGGAAUAUCUUUUAAGCCGCAGUCGGUAACUUGCCGGCUUUAAUGGCAGCGGCUUAGAG -----------------...((((--(....(....(----((((....))))).).))))).-.......-....(((((...)))))((((((((((...((((.....))))(((.....))).)))))))))). ( -30.20, z-score = -1.24, R) >droEre2.scaffold_4690 6237182 112 + 18748788 -----------------CAGCGCU--AAAAACGAAAU----CAGGACAUUCUGAAGGUGGCG--GCAAAGA-AACAAGAUGGAAUAUCUUUUAAGCCGCAGUCGGUAACUUGCCGGCUUUAAUGGCAGCGGCUUAGAG -----------------...((((--(....(....(----((((....))))).).)))))--.......-....(((((...)))))((((((((((...((((.....))))(((.....))).)))))))))). ( -32.20, z-score = -1.35, R) >droAna3.scaffold_13248 2179587 132 - 4840945 UCAUGCCACCCAUGUGCCACCAUUUAAAAAAGAAAAU----CAGAACGAACCGAAGGUGGCGG-CAAAAGA-AACAAGAUGGAAUAUCUUUUAAGCCGUAGUCGGUAACUUGCCGGCUUUAAUGGCAACGGCUUAGAG ...((((((....))((((((.(((......)))..(----(.....))......))))))))-)).....-....(((((...)))))(((((((((((((((((.....))))))).........)))))))))). ( -36.50, z-score = -1.85, R) >dp4.chrXL_group1e 7706159 131 + 12523060 -------UUAUAUCAAAAAUUAGCUGAAAACAGUCAUACCCUCUGUGGGAGGAAGGGUGCCCCAAAGAUGAGAACAAGAUGGAAUAUCUUUUAAGCCAUAGUCGGUAGCUUGCCGGCUUUAAUGGCAGCGGCUUAGAG -------........................((((.((((..((((((...((((((((..(((....((....))...)))..))))))))...))))))..))))(((.((((.......)))))))))))..... ( -34.10, z-score = -0.14, R) >droPer1.super_17 157346 131 + 1930428 -------UUAUAUCAAAAGUUAGCUGAAAAUAGUCAUACCCUCUGUAGGAGGAAGGGUGCCCCAAAGAUGAGAACAAGAUGGAAUAUCUUUUAAGCCAUAGUCGGUAGCUUGCCGGCUUUAAUGGCAGCGGCUUAGAG -------..............(((((.......((((..((((.....))))..(((...)))....))))....((((((...))))))....((((((((((((.....)))))))...)))))..)))))..... ( -33.40, z-score = -0.13, R) >droWil1.scaffold_180588 243322 120 - 1294757 -------ACAUAUUAGAAAAUGUCACAAGAAGGAGGA----CAAAAAAAAGGAAAAAG------AAAAUAA-GACAAGAUGGAAUAUCUUUUAAGCCGUAGUCGGUAACUUGCCGGCUUUAAUGGCAACGGCUUAGAG -------.............((((.(........)))----))...............------.......-....(((((...)))))(((((((((((((((((.....))))))).........)))))))))). ( -24.40, z-score = -1.88, R) >consensus _________________CAGUGCUG_AAAAACGAAAU____CAGGACAUACUGAAGGUGGCGG_AAAAAGA_AACAAGAUGGAAUAUCUUUUAAGCCGCAGUCGGUAACUUGCCGGCUUUAAUGGCAGCGGCUUAGAG .........................................(((......))).......................(((((...)))))(((((((((((((((((.....))))))).........)))))))))). (-22.01 = -21.42 + -0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:45 2011