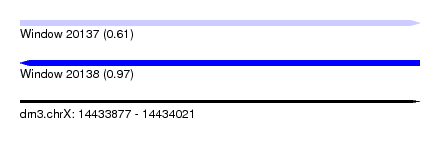
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,433,877 – 14,434,021 |
| Length | 144 |
| Max. P | 0.965013 |

| Location | 14,433,877 – 14,434,021 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 150 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Shannon entropy | 0.34120 |
| G+C content | 0.34023 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -15.21 |
| Energy contribution | -17.18 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14433877 144 + 22422827 UAAAUAAUACAAUCCCAAA-UUGAAAAUACUUAGGCCUUUUAAUGAAUACACUUAUUCAGAGUUUUCUAAUCCCGAACAUCACACCAAUGAUAUAUGAGC-----UGGUGGAACUUUCGUUUCUGGGAUUAGUGGCCUUUAAAGUUUUUC ...................-..(((((.(((((((((......((((((....)))))).......(((((((((.......(((((.............-----)))))((((....)))).))))))))).)))))...))))))))) ( -32.62, z-score = -2.15, R) >droEre2.scaffold_4690 6222626 126 - 18748788 UAAGUAAUAAAAUCCCAA--UUGAAAAUACUUGGCC------------------ACUCGGAGUUUUCUAAUCC-GUACAUCACGCCAGUGAUGUACUCGCCUG---GCUGGAACUUCCAUUUCCGGGAUUAGUGGCCUUUAAAGUUUUCC ..................--..((((((....((((------------------(((.(((.........)))-(((((((((....)))))))))...((((---(.(((.....)))...)))))...)))))))......)))))). ( -41.00, z-score = -4.29, R) >droYak2.chrX 8675745 146 + 21770863 UAAAUAAUAAUAUCUCAAAAUUGAAAAUACUUGGGCCAUUUCCUAAAUA--UUUAUUCACAGUUUUCUAAUCCCGUACAUCACACCAAUGAUAUAUGAGCCAUA--UGAGCAACUUUCGUUUCUGGCAUUAGUAGCCUUUAAAGUUUUGC ...............(((((((..(((((.(((((......))))).))--))).......(((..(((((..((((.((((......)))).)))).((((.(--((((.....)))))...))))))))).)))......))))))). ( -23.20, z-score = -0.67, R) >droSec1.super_20 1031810 149 + 1148123 UAAAUAAUACUAUUCCAAA-UUGAAAAUACUUAGGCCAUUUAAUGAAUACACUUAUUCAGAGUUUUCUAAUCCCGAACAUCACCCCAAUAAUAUAUGAGCAUGACUAGUAGAACUUUCAUUUCUGGGAUUAGUGGCCUUUAAAGUUUUUC ...................-..(((((.((((((((((.....((((((....)))))).......(((((((((.....)...............(((.((((..((.....)).))))))).))))))))))))))...))))))))) ( -29.70, z-score = -2.18, R) >droSim1.chrX 11121587 149 + 17042790 UAAAUAAUACUAUCCCAAA-UUAAAAAUACUUAGGCCAUUUAAUGAAUACACUUAUUCAGAGUUUUCUAAUCCCGAACAUCACACCAAUGAUAUAUGAGCAUGACUGGUAGAACUUUCAUUUCUGGGAUUAGUGGCCUUUAAAGUUUUUC ...................-....(((.((((((((((.....((((((....)))))).......(((((((((...((((......))))....(((.((((..(......)..))))))))))))))))))))))...)))).))). ( -30.60, z-score = -2.07, R) >consensus UAAAUAAUACUAUCCCAAA_UUGAAAAUACUUAGGCCAUUUAAUGAAUACACUUAUUCAGAGUUUUCUAAUCCCGAACAUCACACCAAUGAUAUAUGAGCAUGA_UGGUGGAACUUUCAUUUCUGGGAUUAGUGGCCUUUAAAGUUUUUC ......................((((((....(((((......((((((....)))))).......((((((((....((((......)))).................((((.......)))))))))))).))))).....)))))). (-15.21 = -17.18 + 1.97)

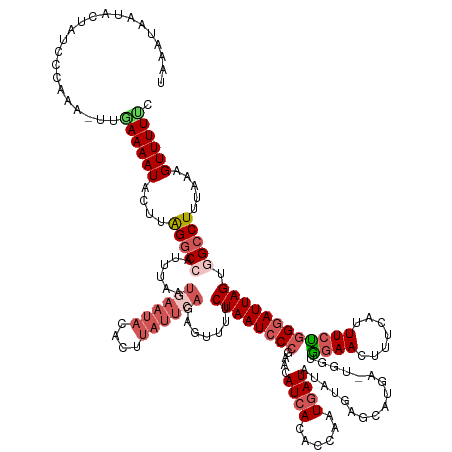
| Location | 14,433,877 – 14,434,021 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 150 |
| Reading direction | reverse |
| Mean pairwise identity | 80.90 |
| Shannon entropy | 0.34120 |
| G+C content | 0.34023 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -24.77 |
| Energy contribution | -25.74 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.965013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
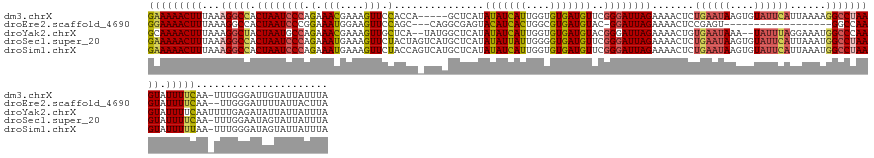
>dm3.chrX 14433877 144 - 22422827 GAAAAACUUUAAAGGCCACUAAUCCCAGAAACGAAAGUUCCACCA-----GCUCAUAUAUCAUUGGUGUGAUGUUCGGGAUUAGAAAACUCUGAAUAAGUGUAUUCAUUAAAAGGCCUAAGUAUUUUCAA-UUUGGGAUUGUAUUAUUUA (((((((((...(((((.((((((((.(((((....))(((((((-----(...........)))))).))..))))))))))).......((((((....))))))......))))))))).)))))..-................... ( -38.50, z-score = -3.23, R) >droEre2.scaffold_4690 6222626 126 + 18748788 GGAAAACUUUAAAGGCCACUAAUCCCGGAAAUGGAAGUUCCAGC---CAGGCGAGUACAUCACUGGCGUGAUGUAC-GGAUUAGAAAACUCCGAGU------------------GGCCAAGUAUUUUCAA--UUGGGAUUUUAUUACUUA (((((((((....(((((((..((((((...(((.....))).)---).)).))(((((((((....)))))))))-(((.........))).)))------------------)))))))).)))))..--.................. ( -39.80, z-score = -2.86, R) >droYak2.chrX 8675745 146 - 21770863 GCAAAACUUUAAAGGCUACUAAUGCCAGAAACGAAAGUUGCUCA--UAUGGCUCAUAUAUCAUUGGUGUGAUGUACGGGAUUAGAAAACUGUGAAUAAA--UAUUUAGGAAAUGGCCCAAGUAUUUUCAAUUUUGAGAUAUUAUUAUUUA .............((((((((((.((.(((((....)))(((..--...))))).((((((((....)))))))).)).))))).....(.(((((...--.))))).)...)))))..((((((((.......))))))))........ ( -28.00, z-score = -0.19, R) >droSec1.super_20 1031810 149 - 1148123 GAAAAACUUUAAAGGCCACUAAUCCCAGAAAUGAAAGUUCUACUAGUCAUGCUCAUAUAUUAUUGGGGUGAUGUUCGGGAUUAGAAAACUCUGAAUAAGUGUAUUCAUUAAAUGGCCUAAGUAUUUUCAA-UUUGGAAUAGUAUUAUUUA (((((((((...((((((((((((((((((.(....)))))....(((((.((((........)))))))))....)))))))).......((((((....)))))).....)))))))))).)))))..-................... ( -38.30, z-score = -2.97, R) >droSim1.chrX 11121587 149 - 17042790 GAAAAACUUUAAAGGCCACUAAUCCCAGAAAUGAAAGUUCUACCAGUCAUGCUCAUAUAUCAUUGGUGUGAUGUUCGGGAUUAGAAAACUCUGAAUAAGUGUAUUCAUUAAAUGGCCUAAGUAUUUUUAA-UUUGGGAUAGUAUUAUUUA (((((((((...(((((((((((((((....)(((.(((.(((((((.(((......))).))))))).))).))))))))))).......((((((....)))))).....)))))))))).)))))..-................... ( -36.30, z-score = -2.29, R) >consensus GAAAAACUUUAAAGGCCACUAAUCCCAGAAAUGAAAGUUCCACCA_UCAGGCUCAUAUAUCAUUGGUGUGAUGUUCGGGAUUAGAAAACUCUGAAUAAGUGUAUUCAUUAAAUGGCCUAAGUAUUUUCAA_UUUGGGAUAGUAUUAUUUA (((((((((...(((((.((((((((.(.(((....))).)...............(((((((....)))))))..)))))))).......((((((....))))))......))))))))).)))))...................... (-24.77 = -25.74 + 0.97)
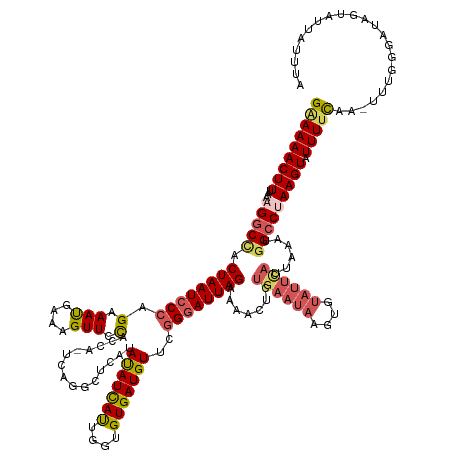
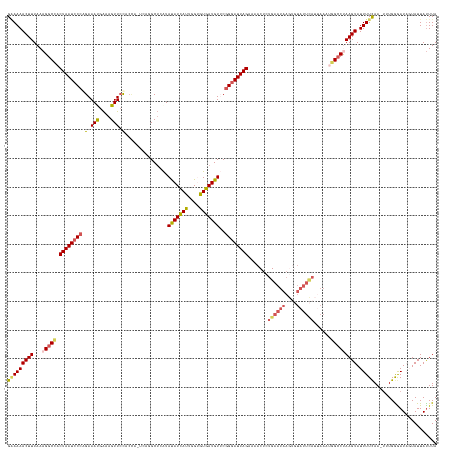
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:42 2011