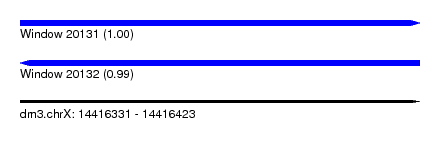
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,416,331 – 14,416,423 |
| Length | 92 |
| Max. P | 0.999717 |

| Location | 14,416,331 – 14,416,423 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 71.41 |
| Shannon entropy | 0.50386 |
| G+C content | 0.49325 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -16.15 |
| Energy contribution | -17.10 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
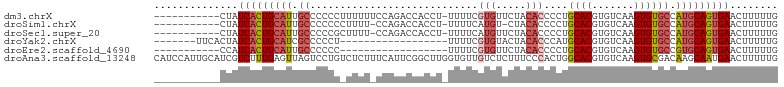
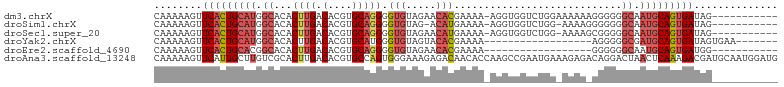
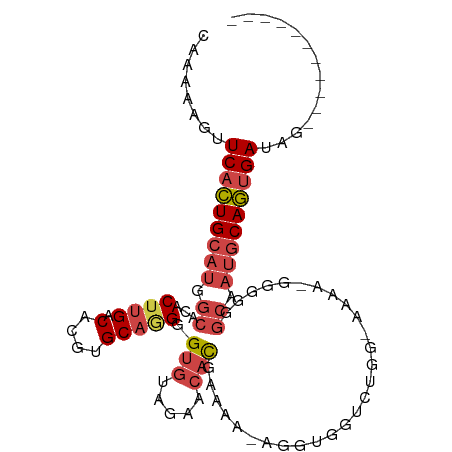
>dm3.chrX 14416331 92 + 22422827 -----------CUAUCACUGCAUUGCCCCCCUUUUUUCCAGACCACCU-UUUUCGUGUUCUACACCCCUGCACGUGUCAAGUGUGCCAUGCAGUGAACUUUUUG -----------...(((((((((.((.............(((.(((..-.....))).)))........((((.......)))))).)))))))))........ ( -23.30, z-score = -4.57, R) >droSim1.chrX 11103511 90 + 17042790 -----------CUAUCACUGCAUUGCCCCCCCUUUU-CCAGACCACCU-UUUUCAUGU-CUACACCCCUGCACGUGUCAAGUGUGCCAUGCAGUGAACUUUUUG -----------...(((((((((.((..........-..((((.....-.......))-))........((((.......)))))).)))))))))........ ( -21.40, z-score = -3.99, R) >droSec1.super_20 1012595 91 + 1148123 -----------CUAUCACUGCAUUGCCCCCGCUUUU-CCAGACCACCU-UUUUCAUGUUCUACACCCCUGCACGUGUCAAGUGUGCCAUGCAGUGAACUUUUUG -----------...(((((((((.((...(((((..-..(((.((...-......)).)))((((........)))).))))).)).)))))))))........ ( -21.20, z-score = -3.29, R) >droYak2.chrX 8657188 79 + 21770863 -------UUCACUAUCACUGCAUCGCCCCCU------------------UUUUCGUGUACUACACCCAUGCACGUGUCAAGUGUGCCAUGCAGUGAACUUUUUG -------.......(((((((((.((.(.((------------------(...((((((.........))))))....))).).)).)))))))))........ ( -23.60, z-score = -4.32, R) >droEre2.scaffold_4690 6206638 75 - 18748788 -----------CCAUCACUGCAUUGCCCCCC------------------UUUUCGUGUUCUACACCCCUGCACGUGUCAAGUGUGCCGUGCAGUGAACUUUUUG -----------...(((((((((.((.....------------------.....(((.....)))....((((.......)))))).)))))))))........ ( -22.10, z-score = -4.10, R) >droAna3.scaffold_13248 2144450 104 + 4840945 CAUCCAUUGCAUCGUCUUUGAGUUAGUCCUGUCUCUUUCAUUCGGCUUGGUGUUGUCUCUUUCCCACUGGCACGUGUCAAGUGCGACAAGCAAUGAACUUUUUG ....((((((........((((..((......))..)))).(((((((((..(((((...........)))).)..)))))).)))...))))))......... ( -22.70, z-score = -0.51, R) >consensus ___________CUAUCACUGCAUUGCCCCCC_UUUU_CCAGACCACCU_UUUUCGUGUUCUACACCCCUGCACGUGUCAAGUGUGCCAUGCAGUGAACUUUUUG ..............(((((((((.((............................(((.....)))....((((.......)))))).)))))))))........ (-16.15 = -17.10 + 0.95)
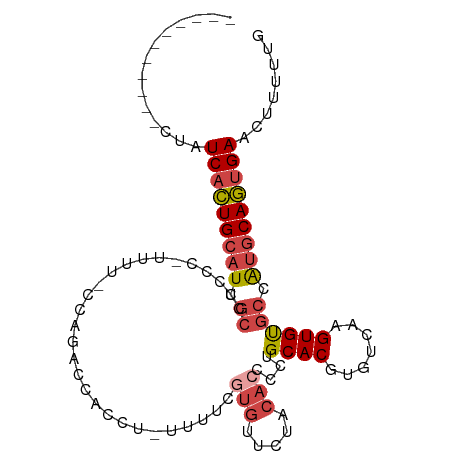
| Location | 14,416,331 – 14,416,423 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 71.41 |
| Shannon entropy | 0.50386 |
| G+C content | 0.49325 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -15.26 |
| Energy contribution | -16.32 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14416331 92 - 22422827 CAAAAAGUUCACUGCAUGGCACACUUGACACGUGCAGGGGUGUAGAACACGAAAA-AGGUGGUCUGGAAAAAAGGGGGGCAAUGCAGUGAUAG----------- ........(((((((((.((...(((.((((........))))(((.(((.....-..))).))).......)))...)).)))))))))...----------- ( -25.80, z-score = -2.68, R) >droSim1.chrX 11103511 90 - 17042790 CAAAAAGUUCACUGCAUGGCACACUUGACACGUGCAGGGGUGUAG-ACAUGAAAA-AGGUGGUCUGG-AAAAGGGGGGGCAAUGCAGUGAUAG----------- ........(((((((((.((...(((.((((........))))((-((.......-.....))))..-....)))...)).)))))))))...----------- ( -23.70, z-score = -1.76, R) >droSec1.super_20 1012595 91 - 1148123 CAAAAAGUUCACUGCAUGGCACACUUGACACGUGCAGGGGUGUAGAACAUGAAAA-AGGUGGUCUGG-AAAAGCGGGGGCAAUGCAGUGAUAG----------- ........(((((((((.((((.((((.(....))))).))))............-...((.(((.(-.....).))).)))))))))))...----------- ( -25.20, z-score = -1.94, R) >droYak2.chrX 8657188 79 - 21770863 CAAAAAGUUCACUGCAUGGCACACUUGACACGUGCAUGGGUGUAGUACACGAAAA------------------AGGGGGCGAUGCAGUGAUAGUGAA------- ........(((((((((.((...(((....((((.((.......)).))))...)------------------))...)).))))))))).......------- ( -25.00, z-score = -2.51, R) >droEre2.scaffold_4690 6206638 75 + 18748788 CAAAAAGUUCACUGCACGGCACACUUGACACGUGCAGGGGUGUAGAACACGAAAA------------------GGGGGGCAAUGCAGUGAUGG----------- ........((((((((..((...((((.(....))))).(((.....))).....------------------.....))..))))))))...----------- ( -21.10, z-score = -1.79, R) >droAna3.scaffold_13248 2144450 104 - 4840945 CAAAAAGUUCAUUGCUUGUCGCACUUGACACGUGCCAGUGGGAAAGAGACAACACCAAGCCGAAUGAAAGAGACAGGACUAACUCAAAGACGAUGCAAUGGAUG ......((((((((((((((..........((.((...(((.............))).)))).......(((..........)))...))))).))))))))). ( -23.82, z-score = -0.97, R) >consensus CAAAAAGUUCACUGCAUGGCACACUUGACACGUGCAGGGGUGUAGAACACGAAAA_AGGUGGUCUGG_AAAA_GGGGGGCAAUGCAGUGAUAG___________ ........(((((((((.((...((((.(....))))).(((.....)))............................)).))))))))).............. (-15.26 = -16.32 + 1.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:36 2011