| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,412,360 – 14,412,459 |
| Length | 99 |
| Max. P | 0.971600 |

| Location | 14,412,360 – 14,412,459 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 63.44 |
| Shannon entropy | 0.69565 |
| G+C content | 0.48965 |
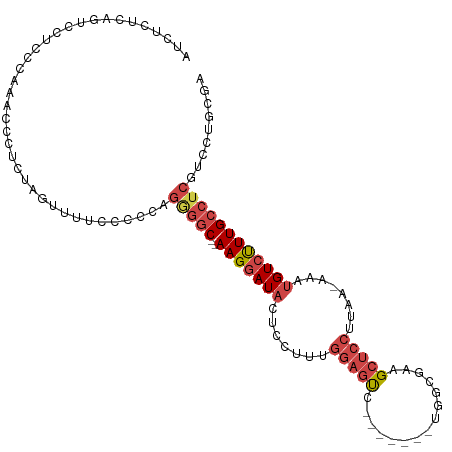
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -12.38 |
| Energy contribution | -13.52 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14412360 99 + 22422827 AUCUCUCAGUCCCC---AACCCUCUAGUUGUCCGCCAGGGGC--AAGGAUACUCCUUUGGAGUC-------UGGCGAAGCUCCUUAA-AAAUGUCUUUGCCUCGUCCUGCAA ......(((..(.(---(((......)))).......(((((--(((((((.......((((.(-------(.....))))))....-...)))))))))))))..)))... ( -29.04, z-score = -1.80, R) >droAna3.scaffold_13248 2140994 98 + 4840945 -UAUAUUUUUCCUCAAAAAACUUUCAGCGAAAAGCAAAGUUCUCAAGAAUCAAGAAUUAAUAUCCUCUGUCUAUUUUAGAUCAAAAAUAAAUGUCCUUG------------- -..(((((((..((....((((((..((.....))))))))....(((....(((..........))).)))......))..)))))))..........------------- ( -8.40, z-score = 0.09, R) >droEre2.scaffold_4690 6202875 103 - 18748788 AUCCCUCACUCCGC---AACCCUUUGCUUUUCCCC--GAGGC--AAGGAUACUCCUUUGGAGUCGGAGUC-UGGCGAAGCUCCUUAA-AAAUGUCCUUGCCUCCUCCUGCAA ............((---((....))))........--(((((--(((((((((((...))))..((((.(-(.....))))))....-...))))))))))))......... ( -33.70, z-score = -3.37, R) >droYak2.chrX 8653290 101 + 21770863 --------AUCUCCCCAACCUCUGUAGUUGUUCCCCCGAGGC--AAGGAUACUCCUCUGGAGCUGGACUCCUAAGGAAGCUCCUUAA-AAAUGUCUUUGCCUCCUCCUGCGA --------..............(((((..(.......(((((--(((((((.......((((((...((....))..))))))....-...)))))))))))))..))))). ( -30.75, z-score = -2.58, R) >droSec1.super_20 1007533 102 + 1148123 AUCUCUCGGUCCUCCCAAUCCCCCAAGUUUUCCCCCAGGGGC--AAGGAUACAUCUUUGGAGUC-------UGGCGAAGCUCCUUAA-AAAUGUCUUUGCCUCGUCCUGCGA .......((.....)).....................(((((--(((((((.......((((.(-------(.....))))))....-...))))))))))))......... ( -25.84, z-score = -0.25, R) >droSim1.chrX 11099523 102 + 17042790 AUCUCUCGGUCCUCCCAAUCCCCCAAGUUUUCCCCCAGGGGC--AAGGAUACUCCUUGGGAGUC-------UGGCGAAGCUCCUUAA-AAAUGUCUUUGCCUCGUCCUGCGA .......((.((((((....((((.............)))).--((((.....)))))))))..-------.((((((((.......-....).)))))))..).))..... ( -28.32, z-score = -0.35, R) >consensus AUCUCUCAGUCCUCCCAAACCCUCUAGUUUUCCCCCAGGGGC__AAGGAUACUCCUUUGGAGUC_______UGGCGAAGCUCCUUAA_AAAUGUCUUUGCCUCGUCCUGCGA .....................................(((((..(((((((.......(((((...............)))))........))))))))))))......... (-12.38 = -13.52 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:35 2011