| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,405,756 – 14,405,847 |
| Length | 91 |
| Max. P | 0.763441 |

| Location | 14,405,756 – 14,405,847 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 72.43 |
| Shannon entropy | 0.51103 |
| G+C content | 0.51249 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -10.38 |
| Energy contribution | -10.55 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14405756 91 + 22422827 ACACUUGGCCUCGGAUCGGAUCCGAGCA----GCAGGACUCCGGUUGUGGAAAUAACGAAGACAAAGGCCGACAACAUCGACUACAUACAGACAG ....(((((((..(..(((((((.....----...))).))))(((((....))))).....)..)))))))....................... ( -24.90, z-score = -1.82, R) >droSim1.chrX 11092841 91 + 17042790 ACACUUGGCCUCGGAUUGGAUCAGAGCA----GCAGGACGCCGGGUGUGGAAAUAACGAAGACAAAGGCCGACAACAUCGGCGACAUACACACAU (((((((((.((.(.(((........))----))..)).)))))))))...................(((((.....)))))............. ( -22.80, z-score = -1.02, R) >droSec1.super_20 999483 91 + 1148123 ACACUUGGCCUCGGAUUGGAUCAGAGCA----GCAGGACGCCGGGUGUGGAAAUAACGAAGACAAAGGCCGACAACAUCGGCGACAUACACACAU (((((((((.((.(.(((........))----))..)).)))))))))...................(((((.....)))))............. ( -22.80, z-score = -1.02, R) >droYak2.chrX 8644483 87 + 21770863 ACACUUGGCCUCGCAUUGGAUGAGAGGACGAAACAGGAUUCCGGGUGUGGAAAUAACGAAGACAAAGGCCGACAACAUCGGCGACAU-------- ((((((((((((.(((...))).)))).(......)....))))))))...................(((((.....))))).....-------- ( -22.40, z-score = -1.11, R) >droWil1.scaffold_180588 194383 81 + 1294757 ACGCUCGCUCACUCACUUGGCUAGACAG------AUGAGCGUGCGAGUGAAAAUAACGAAGACAAAAGCCGACAACAUUGGCGACAU-------- .(((((((.(.((((.(((......)))------.)))).).)))))))..................(((((.....))))).....-------- ( -26.90, z-score = -3.60, R) >droGri2.scaffold_15203 2975760 74 - 11997470 ACACUUGGCCACAC-UUGGAU--AGACA-----CAGAG---CGAGUGU--AAAUAAGGAAGAUAAAAGCCGACAACACUGGCGACUU-------- ...((((...((((-(((...--.....-----.....---)))))))--...))))..........((((.......)))).....-------- ( -14.02, z-score = -1.27, R) >consensus ACACUUGGCCUCGCAUUGGAUCAGAGCA____GCAGGACGCCGGGUGUGGAAAUAACGAAGACAAAGGCCGACAACAUCGGCGACAU________ ((((((((................................))))))))...................(((((.....)))))............. (-10.38 = -10.55 + 0.17)

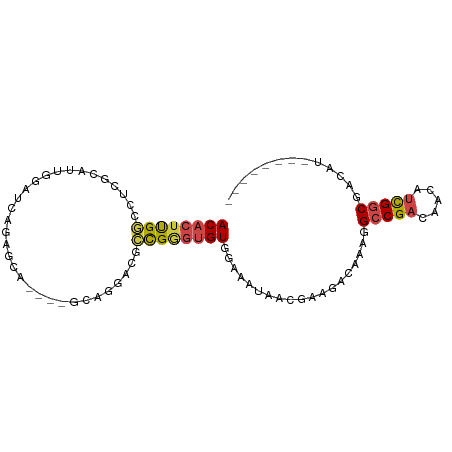

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:34 2011