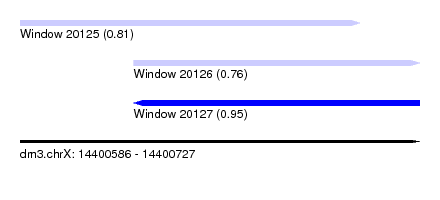
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,400,586 – 14,400,727 |
| Length | 141 |
| Max. P | 0.945431 |

| Location | 14,400,586 – 14,400,706 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.15 |
| Shannon entropy | 0.37648 |
| G+C content | 0.48119 |
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -23.25 |
| Energy contribution | -24.64 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14400586 120 + 22422827 GCAGUUCUGGGCUAAGGGAUAAAGGAUAAAGGACACUGCUUUGGAUUCUCGGCAGGAUUAAUGCCGAAAGGCUUCAUUUGUGAGGCCGGCAAAUGGCAAGUUUUGGCAGAGAAAAGUGUU ((..((((..((((((((((.((((....((....)).))))..))))((((((.......))))))..(((((((....))))))).((.....))....))))))..))))..))... ( -34.80, z-score = -1.03, R) >droSim1.chrX 11087717 120 + 17042790 GCAGUUCUGGGUUAAGGGAUAAAGGAUAAAGGACACUGCCUUGAAUUCUCGGCAGGAUUAAUGCCGAAAGGCUUCAUUUGUGAGGCCGCCAAAUGGCAAGUUUUGGCACAGAAAAGUGGG ((..(((((.((..((((...((((....((....)).))))...))))..))........(((((((((((((((....)))))))(((....)))...)))))))))))))..))... ( -41.70, z-score = -2.83, R) >droSec1.super_20 994372 120 + 1148123 GUAGUUCUGGGUUAAGGGAUAAAGGAUAAAGGACACUGCCUUGGAUUCUCAGCAGGAUUAAUGCCGAAAGGCUUCAUUUGUGAGGCCGCCAAAUGGCAAGUUUUGGCACAGAAAAGUGGU ....(((((......((((..((((....((....)).))))....))))...........(((((((((((((((....)))))))(((....)))...)))))))))))))....... ( -39.10, z-score = -2.26, R) >droYak2.chrX 8639345 112 + 21770863 GCGCUUCUGGGUUAAGGGAUAAAGGAUAAAGGACACCGGCUUCGAUUCUCGGCAGGAUUAAUGCCAAAAGGCUUCACUUGUGUA------AAGUGGCAAGUUUUGGCACAGAAGUGUU-- (((((((((......((((....(((....((...))...)))...))))...........((((((((.((..(((((.....------)))))))...))))))))))))))))).-- ( -32.50, z-score = -1.22, R) >droEre2.scaffold_4690 6192938 96 - 18748788 GCGGCGCUGGGUUAAGGGAUACAGGAUAAAGGACGCUGCCUCUGAUUCUCGCCGGGAUUAAGGCCAACA------------------------UGGCAGGUUUCGGCACAGAAAAGUGAU ((((((((..(((...(....)..)))...)).))))))(.((..((((.((((..(((...(((....------------------------.))).)))..))))..)))).)).).. ( -31.20, z-score = -1.13, R) >consensus GCAGUUCUGGGUUAAGGGAUAAAGGAUAAAGGACACUGCCUUGGAUUCUCGGCAGGAUUAAUGCCGAAAGGCUUCAUUUGUGAGGCCG_CAAAUGGCAAGUUUUGGCACAGAAAAGUGGU ....(((((.((...((((..((((....((....)).))))....))))..(((((((..(((((...(((((((....)))))))......)))))))))))))).)))))....... (-23.25 = -24.64 + 1.39)

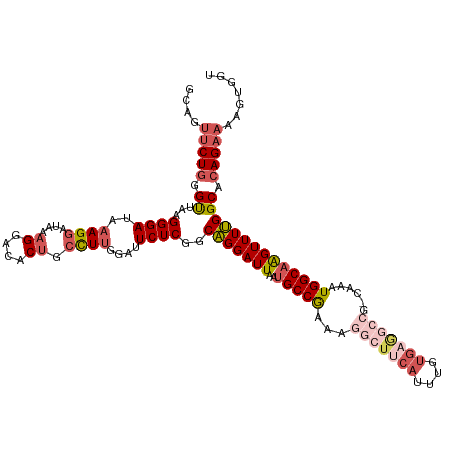
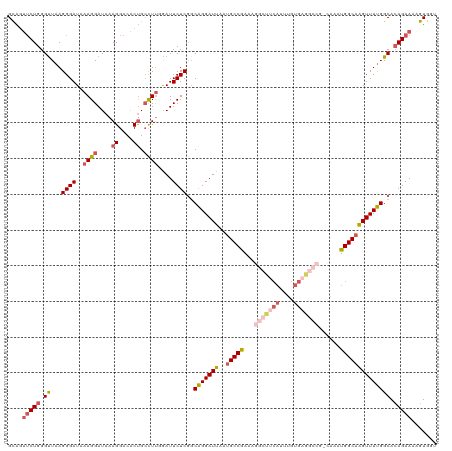
| Location | 14,400,626 – 14,400,727 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Shannon entropy | 0.36127 |
| G+C content | 0.41578 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -18.44 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.762852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14400626 101 + 22422827 UUGGAUUCUCGGCAGGAUUAAUGCCGAAAGGCUUCAUUUGUGAGGCCGGCAAAUGGCAAGUUUUGGCAGAGAAAAGUGUUGAAAGAUAUGGUUUAGUUUUA----------------- .....((((((.(((((((..(((((...(((((((....)))))))......)))))))))))).).)))))..(((((....)))))............----------------- ( -30.20, z-score = -2.47, R) >droSim1.chrX 11087757 101 + 17042790 UUGAAUUCUCGGCAGGAUUAAUGCCGAAAGGCUUCAUUUGUGAGGCCGCCAAAUGGCAAGUUUUGGCACAGAAAAGUGGGGAAAGAUAUGGCUUAGUUUUA----------------- .....(((((.((........(((((((((((((((....)))))))(((....)))...)))))))).......)).)))))..................----------------- ( -31.76, z-score = -2.14, R) >droSec1.super_20 994412 101 + 1148123 UUGGAUUCUCAGCAGGAUUAAUGCCGAAAGGCUUCAUUUGUGAGGCCGCCAAAUGGCAAGUUUUGGCACAGAAAAGUGGUGAAAGGUAUGGGCUAGUUUUA----------------- (((((((((....))))))..(((((((((((((((....)))))))(((....)))...)))))))))))((((.((((...........)))).)))).----------------- ( -30.80, z-score = -1.60, R) >droYak2.chrX 8639385 112 + 21770863 UUCGAUUCUCGGCAGGAUUAAUGCCAAAAGGCUUCACUUGUGUA------AAGUGGCAAGUUUUGGCACAGAAGUGUUGAAAAAUAUGGGUUUUAACUCACCAAAAAAAUGAGUUUUA ...((..(((((((.......))))....((..((((((.....------))))))....(((..((((....))))..)))....(((((....)))))))........)))..)). ( -26.10, z-score = -0.97, R) >consensus UUGGAUUCUCGGCAGGAUUAAUGCCGAAAGGCUUCAUUUGUGAGGCCGCCAAAUGGCAAGUUUUGGCACAGAAAAGUGGUGAAAGAUAUGGUUUAGUUUUA_________________ ...((((((....))))))..(((((((((((((((....)))))))(((....)))...)))))))).................................................. (-18.44 = -19.88 + 1.44)

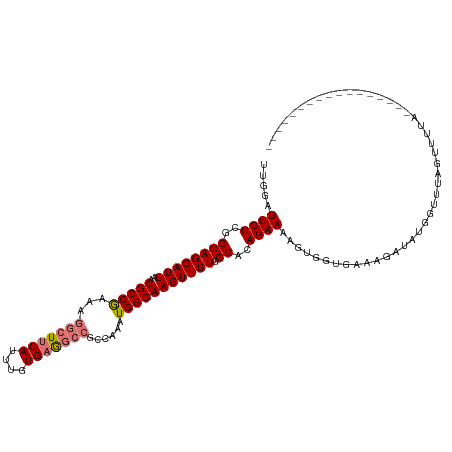
| Location | 14,400,626 – 14,400,727 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Shannon entropy | 0.36127 |
| G+C content | 0.41578 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -13.35 |
| Energy contribution | -12.85 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
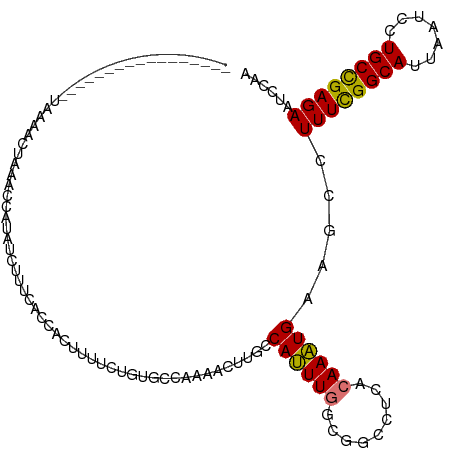
>dm3.chrX 14400626 101 - 22422827 -----------------UAAAACUAAACCAUAUCUUUCAACACUUUUCUCUGCCAAAACUUGCCAUUUGCCGGCCUCACAAAUGAAGCCUUUCGGCAUUAAUCCUGCCGAGAAUCCAA -----------------............................(((((.((.......((((.......(((.(((....))).)))....))))........)).)))))..... ( -16.96, z-score = -2.10, R) >droSim1.chrX 11087757 101 - 17042790 -----------------UAAAACUAAGCCAUAUCUUUCCCCACUUUUCUGUGCCAAAACUUGCCAUUUGGCGGCCUCACAAAUGAAGCCUUUCGGCAUUAAUCCUGCCGAGAAUUCAA -----------------.........(((............((......))((((((........)))))))))........((((...((((((((.......)))))))).)))). ( -24.30, z-score = -2.83, R) >droSec1.super_20 994412 101 - 1148123 -----------------UAAAACUAGCCCAUACCUUUCACCACUUUUCUGUGCCAAAACUUGCCAUUUGGCGGCCUCACAAAUGAAGCCUUUCGGCAUUAAUCCUGCUGAGAAUCCAA -----------------.................(((((.((.......(((((.......(((....)))(((.(((....))).)))....)))))......)).)))))...... ( -22.12, z-score = -2.05, R) >droYak2.chrX 8639385 112 - 21770863 UAAAACUCAUUUUUUUGGUGAGUUAAAACCCAUAUUUUUCAACACUUCUGUGCCAAAACUUGCCACUU------UACACAAGUGAAGCCUUUUGGCAUUAAUCCUGCCGAGAAUCGAA ...((((((((.....))))))))...........(((((..((.....(((((((((...(((((((------.....)))))..)).)))))))))......))..)))))..... ( -23.60, z-score = -2.26, R) >consensus _________________UAAAACUAAACCAUAUCUUUCACCACUUUUCUGUGCCAAAACUUGCCAUUUGGCGGCCUCACAAAUGAAGCCUUUCGGCAUUAAUCCUGCCGAGAAUCCAA ...............................................................((((((.........)))))).....((((((((.......))))))))...... (-13.35 = -12.85 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:32 2011