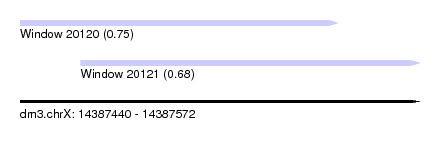
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,387,440 – 14,387,572 |
| Length | 132 |
| Max. P | 0.748358 |

| Location | 14,387,440 – 14,387,545 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 70.15 |
| Shannon entropy | 0.53713 |
| G+C content | 0.36195 |
| Mean single sequence MFE | -20.36 |
| Consensus MFE | -11.86 |
| Energy contribution | -13.18 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14387440 105 + 22422827 --UUCAUACAAAUUAAU--UCGAAAUUUUAGAACUAUAUUACUAAUCGCCAUUCACACGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGAUUGUUACCAUUGCAUUU --...............--(((.((((((((..........))).(((((...(((((.((((....)))).))))).)))))))))).)))................. ( -17.30, z-score = -0.22, R) >droYak2.chrX 8626398 107 + 21770863 --UUCGUACAUAUAAAAAAUUCAAAUUUCAGCACUAGUUUACUAAGCGCCAUUCACAAGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGACUGUUACCAUUGCAUUU --.(((((................(((((.....(((....)))..((((...((((..((((....))))..)))).))))))))))))))................. ( -18.19, z-score = 0.14, R) >droSec1.super_20 981665 105 + 1148123 --UUUGUACAAAUGAAU--UCGAAAUUUCAGCACUAGUUUACUAAUCGCCAUUCACACGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGAUUGUUACCAUAGCAUUU --..(((.....(((..--(((.((((((.....(((....)))..((((...(((((.((((....)))).))))).)))))))))).)))...))).....)))... ( -23.40, z-score = -1.29, R) >droSim1.chrX 11074639 105 + 17042790 --UUGGUACAAAUGAAU--UCGAAAAUUCAGCACUAGUUUACUAAUCGCCAUUCACACGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGAUUGUUACCAUUGCAUUU --.((((((((.(((((--(....))))))......((..(((..(((((...(((((.((((....)))).))))).))))).))).))..))).)))))........ ( -26.00, z-score = -1.74, R) >droVir3.scaffold_13049 2747543 89 - 25233164 GAUGUAUA-AAACAAGUCGGAUUGGUUACAUCUCUGGCUCACCAA--AUCUUCCACA-AUAAAACUUUAAUUAU-UGUGUUCGACAUUACGCCU--------------- ........-......(((((((((((..((....))....)))))--......((((-((((........))))-)))))))))).........--------------- ( -16.90, z-score = -2.01, R) >consensus __UUCGUACAAAUAAAU__UCGAAAUUUCAGCACUAGUUUACUAAUCGCCAUUCACACGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGAUUGUUACCAUUGCAUUU ...................(((.((((((.....(((....)))..((((...(((((.((((....)))).))))).)))))))))).)))................. (-11.86 = -13.18 + 1.32)



| Location | 14,387,460 – 14,387,572 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.64 |
| Shannon entropy | 0.53262 |
| G+C content | 0.42629 |
| Mean single sequence MFE | -26.81 |
| Consensus MFE | -13.56 |
| Energy contribution | -16.48 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14387460 112 + 22422827 -AUUUUAGAACUAUAUUACUAAUCGCCAUUCACACGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGAUUGUUACCAUUGCAUUUUGGCGGUGGCCAAUUGGUUUUUAGCCG -.....(((((...........(((((...(((((.((((....)))).))))).)))))......((((((...(((((((......))))))).)))))))))))...... ( -31.00, z-score = -1.85, R) >droEre2.scaffold_4690 6180392 89 - 18748788 -AUUUCAGCGCUAGUUUACUAAUCGCCACUCACA-----------------------CGGAAGUUCCGACUGUUACCAUUGCAUUUUGGCGGUGGCCAAUUGGUUUUUAGCCG -......(.(((((...((((((.((((((....-----------------------(((.....))).......(((........))).))))))..))))))..)))))). ( -24.80, z-score = -1.90, R) >droYak2.chrX 8626420 112 + 21770863 -AUUUCAGCACUAGUUUACUAAGCGCCAUUCACAAGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGACUGUUACCAUUGCAUUUUGGCGGUGGCCAAUUGGUUUUUAGCUG -....((((..(((((.(((...((((...((((..((((....))))..)))).))))..)))...)))))...(((((((......)))))))..............)))) ( -30.80, z-score = -0.93, R) >droSec1.super_20 981685 112 + 1148123 -AUUUCAGCACUAGUUUACUAAUCGCCAUUCACACGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGAUUGUUACCAUAGCAUUUUGGCGGUGGCCAAUUGGUUUUUAGCCG -......((.............(((((...(((((.((((....)))).))))).)))))......((((((...((((.((......)).)))).)))))).......)).. ( -28.30, z-score = -0.69, R) >droSim1.chrX 11074659 112 + 17042790 -AAUUCAGCACUAGUUUACUAAUCGCCAUUCACACGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGAUUGUUACCAUUGCAUUUUGGCGGUGGCCAAUUGGUUUUUAGCCG -......((.............(((((...(((((.((((....)))).))))).)))))......((((((...(((((((......))))))).)))))).......)).. ( -31.30, z-score = -1.42, R) >droVir3.scaffold_13049 2747565 91 - 25233164 GGUUACAUCUCUGGCUCACCAA--AUCUUCCACA-----------------AUAAAACUUUAAUUAUUGUGUUCGACAUUACGCCUUAGAAAUUG---AUCGGGAUUUUGCAA (((((..(((..(((.......--.((...((((-----------------((((........))))))))...))......)))..)))...))---)))............ ( -14.64, z-score = -0.69, R) >consensus _AUUUCAGCACUAGUUUACUAAUCGCCAUUCACACGUUCAAUUAUGGAAGUGUGCGGCGGAAGUUACGAUUGUUACCAUUGCAUUUUGGCGGUGGCCAAUUGGUUUUUAGCCG .(((((.....(((....)))..((((...(((((.((((....)))).))))).)))))))))..((((((...(((((((......))))))).))))))........... (-13.56 = -16.48 + 2.92)

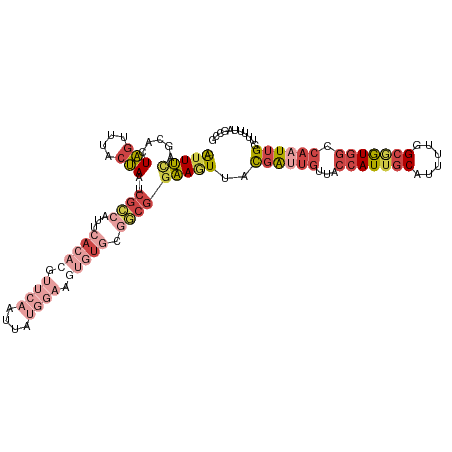

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:28 2011