| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,375,900 – 14,376,009 |
| Length | 109 |
| Max. P | 0.998872 |

| Location | 14,375,900 – 14,376,009 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 65.10 |
| Shannon entropy | 0.61922 |
| G+C content | 0.51506 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -12.07 |
| Energy contribution | -13.60 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.998872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
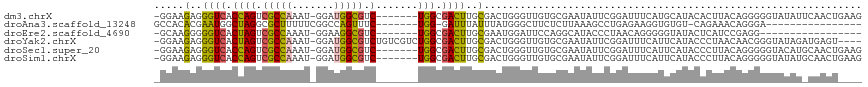
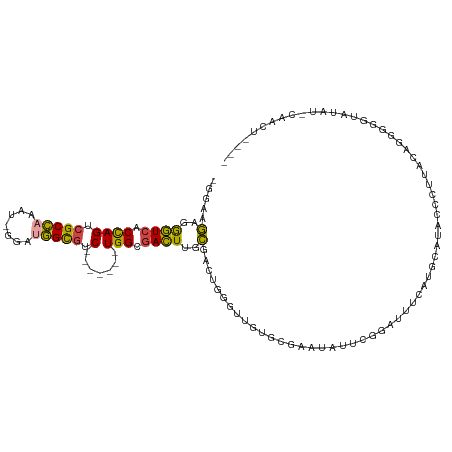
>dm3.chrX 14375900 109 + 22422827 CUUCAGUUGAAUAUACCCCCUGUAAGUGUAUGCAUGAAAUCCGAAUAUUCGCACAACCCAGUCGCAAGUCGCCA-------GACGCCAUCC-AUUUGGCGACUGGUGACCCUCUUCC- .((((.....((((((.........))))))...))))............((((......)).))..(((((((-------(.(((((...-...))))).))))))))........- ( -30.00, z-score = -3.03, R) >droAna3.scaffold_13248 2096204 93 + 4840945 ----------------UCCCUGUUUCUG-ACACACCUUCUCAGGCUUUAAGAGAAGCCCAUAAAUAAAUC-CCA-------GAAACUGGCCGAAAAAGCGCCUAGCCAUUCGUGUGGC ----------------.....(((((((-.............((((((....))))))............-.))-------))))).((((......).)))..(((((....))))) ( -23.97, z-score = -2.03, R) >droEre2.scaffold_4690 6166590 92 - 18748788 -----------------CCUCGGAUGAGUAUACCCCCUGUUAGGGUAUGCCUGGAAUCCAUUCGCAAGUCGCCA-------GACGCCUUCC-AUUUGGCGACUAGUGACCCCCUUGC- -----------------....((((..((((((((.......)))))))).....))))..((((.((((((((-------((.(.....)-.)))))))))).)))).........- ( -34.50, z-score = -3.74, R) >droYak2.chrX 8614060 112 + 21770863 ----ACUCAUCUAUACCCGUUGUUAGGGUAUGAAUGAAAUCCGAAUAUUCGCACAACCCAGUCGCAAGUCGCCAGACGACAGACGCCAUCC-AUUUGGCGACUAGUGACCCUCUUCC- ----..((((.(((((((.......))))))).))))....(((....))).........(((((.((((((((((.(............)-.)))))))))).)))))........- ( -34.90, z-score = -3.86, R) >droSec1.super_20 970364 109 + 1148123 CUUCAGUUGCAUGUACCCCCUGUAAGGGUAUGAAUGAAAUCCGAAUAUUCGCACAACCCAGUCGCAAGUCGCCA-------GACGCCAUCC-AUUUGGCGACUGGUGACCCUCUUCC- ......((((..((((((.......))))))(((((.........))))).............))))(((((((-------(.(((((...-...))))).))))))))........- ( -34.90, z-score = -3.28, R) >droSim1.chrX 11063237 109 + 17042790 CUUCAGUUGCAUAUACCCCCUGUAAGGGUAUGAAUGAAAUCCGAAUAUUCGCACAACCCAGUCGCAAGUCGCCA-------GACGCCAUCC-AUUUGGCGACUGGUGACCCUCUUCC- ......((((.(((((((.......))))))).........(((....)))............))))(((((((-------(.(((((...-...))))).))))))))........- ( -35.50, z-score = -3.88, R) >consensus ____AGUUG_AUAUACCCCCUGUAAGGGUAUGAAUGAAAUCCGAAUAUUCGCACAACCCAGUCGCAAGUCGCCA_______GACGCCAUCC_AUUUGGCGACUAGUGACCCUCUUCC_ ............................................................(((((.((((((((.....................)))))))).)))))......... (-12.07 = -13.60 + 1.53)


| Location | 14,375,900 – 14,376,009 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 65.10 |
| Shannon entropy | 0.61922 |
| G+C content | 0.51506 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -12.18 |
| Energy contribution | -11.49 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.882502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14375900 109 - 22422827 -GGAAGAGGGUCACCAGUCGCCAAAU-GGAUGGCGUC-------UGGCGACUUGCGACUGGGUUGUGCGAAUAUUCGGAUUUCAUGCAUACACUUACAGGGGGUAUAUUCAACUGAAG -....(..((((.((((.(((((...-...))))).)-------))).))))..)...(((((((((((((.........)))..))))).)))))(((..((.....))..)))... ( -34.10, z-score = -1.57, R) >droAna3.scaffold_13248 2096204 93 - 4840945 GCCACACGAAUGGCUAGGCGCUUUUUCGGCCAGUUUC-------UGG-GAUUUAUUUAUGGGCUUCUCUUAAAGCCUGAGAAGGUGUGU-CAGAAACAGGGA---------------- ((((......))))..(((.(......)))).(((((-------(((-.((...(((.(((((((......))))))).)))...)).)-))))))).....---------------- ( -29.80, z-score = -1.17, R) >droEre2.scaffold_4690 6166590 92 + 18748788 -GCAAGGGGGUCACUAGUCGCCAAAU-GGAAGGCGUC-------UGGCGACUUGCGAAUGGAUUCCAGGCAUACCCUAACAGGGGGUAUACUCAUCCGAGG----------------- -((...((((((...((((((((.((-(.....))).-------))))))))........))))))..))(((((((.....))))))).(((....))).----------------- ( -35.90, z-score = -2.56, R) >droYak2.chrX 8614060 112 - 21770863 -GGAAGAGGGUCACUAGUCGCCAAAU-GGAUGGCGUCUGUCGUCUGGCGACUUGCGACUGGGUUGUGCGAAUAUUCGGAUUUCAUUCAUACCCUAACAACGGGUAUAGAUGAGU---- -.......((((.(.((((((((...-.((((((....)))))))))))))).).)))).....((.(((....))).))(((((..((((((.......))))))..))))).---- ( -40.00, z-score = -2.37, R) >droSec1.super_20 970364 109 - 1148123 -GGAAGAGGGUCACCAGUCGCCAAAU-GGAUGGCGUC-------UGGCGACUUGCGACUGGGUUGUGCGAAUAUUCGGAUUUCAUUCAUACCCUUACAGGGGGUACAUGCAACUGAAG -((..(..((((.((((.(((((...-...))))).)-------))).))))..)..)).((((((.(((....)))...........((((((.....))))))...)))))).... ( -38.90, z-score = -2.23, R) >droSim1.chrX 11063237 109 - 17042790 -GGAAGAGGGUCACCAGUCGCCAAAU-GGAUGGCGUC-------UGGCGACUUGCGACUGGGUUGUGCGAAUAUUCGGAUUUCAUUCAUACCCUUACAGGGGGUAUAUGCAACUGAAG -((..(..((((.((((.(((((...-...))))).)-------))).))))..)..)).((((((.(((....)))..........(((((((.....)))))))..)))))).... ( -40.20, z-score = -2.75, R) >consensus _GGAAGAGGGUCACCAGUCGCCAAAU_GGAUGGCGUC_______UGGCGACUUGCGACUGGGUUGUGCGAAUAUUCGGAUUUCAUGCAUACCCUUACAGGGGGUAUAU_CAACU____ .....(..((((.((((.(((((.......))))).).......))).))))..)............................................................... (-12.18 = -11.49 + -0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:23 2011