| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,336,034 – 14,336,171 |
| Length | 137 |
| Max. P | 0.975160 |

| Location | 14,336,034 – 14,336,171 |
|---|---|
| Length | 137 |
| Sequences | 5 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 69.05 |
| Shannon entropy | 0.55860 |
| G+C content | 0.43492 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -14.90 |
| Energy contribution | -17.14 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
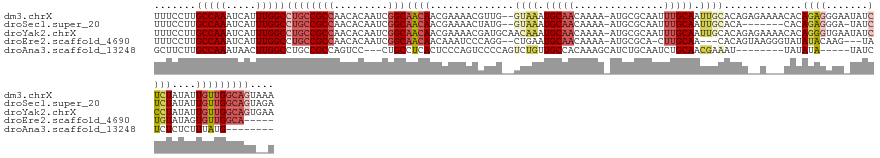
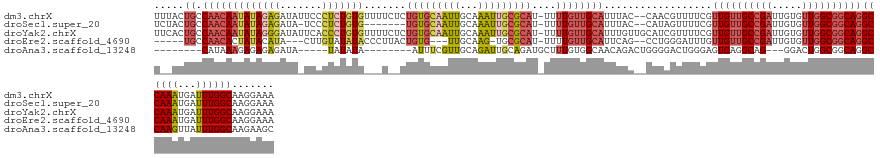
>dm3.chrX 14336034 137 + 22422827 UUUCCUUGCCAAAUCAUUUGGCCUGCCGCCAACACAAUCGGCAACAACGAAAACGUUG--GUAAAUGCAACAAAA-AUGCGCAAUUUGCAAUUGCACAGAGAAAACACAGAGGGAAUAUCUCUAUAUUGUUGGCAGUAAA .......(((((.....))))).(((.(((((((...((.(((((((((....)))))--((((((((.......-....)).))))))..))))...))........(((((.....)))))....))))))).))).. ( -36.40, z-score = -1.93, R) >droSec1.super_20 931245 129 + 1148123 UUUCCUUGCCAAAUCAUUUGGCCUGCCGCCAACACAAUCGGCAACAACGAAAACUAUG--GUAAAUGCAACAAAA-AUGCGCAAUUUGCAAUUGCACA-------CACAGAGGGA-UAUCUCUAUAUUGUUGGCAGUAGA .......(((((.....)))))((((.(((((((...(((.......)))......((--((((.(((((.....-.........))))).)))).))-------...(((((..-..)))))....))))))).)))). ( -33.24, z-score = -1.48, R) >droYak2.chrX 8573232 139 + 21770863 UUUCCUUGCCAAAUCAUUUGGCCUGCCGCCAACACAAUCGGCAACAACGAAAACGAUGCAACAAAUGCAACAAAA-AUGCGCAAUUUGCAAUUGCACAGAGAAAACACAGGGUGAAUAUCCCUAUAUUGUUGGCAGUGAA .......(((((.....)))))((((((((.........)))...(((((...(..(((((.....(((......-.)))((.....))..)))))..).........((((.......))))...)))))))))).... ( -33.20, z-score = -0.56, R) >droEre2.scaffold_4690 6131537 125 - 18748788 UUUCCUUGCCAAAUCAUUUGGCCUGCCGCCAACACAAUCGGCAACAACAAAUCCCAGG--CUGAAUGCAACAAAA-AUGCGCA-CUUGCAA---CACAGUAAGGGUAUAUACAAG---UAUGUAUAGUGUUGGCA----- .......(((((.....))))).....((((((((....(....).......(((..(--(((...(((......-.)))((.-...))..---..))))..)))..((((((..---..)))))))))))))).----- ( -38.10, z-score = -2.28, R) >droAna3.scaffold_13248 2038818 116 + 4840945 GCUUCUUGCCAAAUAACUUGGCCUGCCGCCAGUCC---CUGCCUCACUCCCAGUCCCCAGUCUGUUGCCACAAAGCAUCUGCAAUCUGCAACGAAAU--------UAUAUA-----UAUCUCUCUCUUUAUG-------- ((((..((.(((.....(((((.....)))))...---............(((.(....).)))))).))..))))...(((.....))).......--------......-----................-------- ( -14.90, z-score = 0.14, R) >consensus UUUCCUUGCCAAAUCAUUUGGCCUGCCGCCAACACAAUCGGCAACAACGAAAACCAUG__GUAAAUGCAACAAAA_AUGCGCAAUUUGCAAUUGCACAG__A___CACAGAGGGA_UAUCUCUAUAUUGUUGGCAGU__A .......(((((.....))))).....(((((((..........................((((.(((((...............))))).)))).............(((((.....)))))....)))))))...... (-14.90 = -17.14 + 2.24)
| Location | 14,336,034 – 14,336,171 |
|---|---|
| Length | 137 |
| Sequences | 5 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 69.05 |
| Shannon entropy | 0.55860 |
| G+C content | 0.43492 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -22.77 |
| Energy contribution | -25.14 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
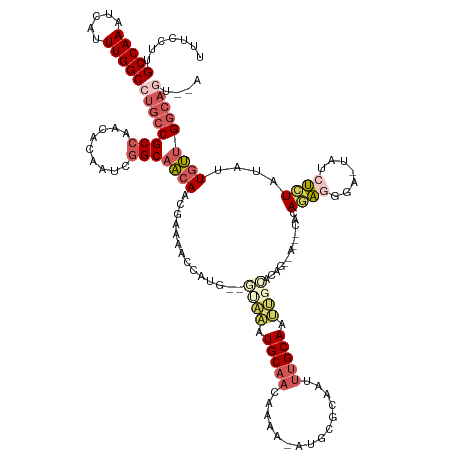
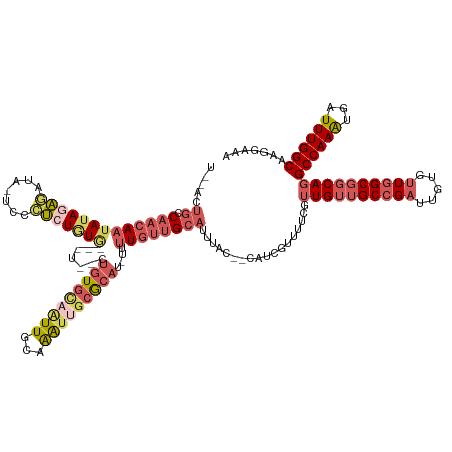
>dm3.chrX 14336034 137 - 22422827 UUUACUGCCAACAAUAUAGAGAUAUUCCCUCUGUGUUUUCUCUGUGCAAUUGCAAAUUGCGCAU-UUUUGUUGCAUUUAC--CAACGUUUUCGUUGUUGCCGAUUGUGUUGGCGGCAGGCCAAAUGAUUUGGCAAGGAAA ....(((((((((((((((((.......)))))))))........((((((((((..((((((.-...)).)))).....--(((((....)))))))).)))))))))))))))...(((((.....)))))....... ( -43.40, z-score = -2.71, R) >droSec1.super_20 931245 129 - 1148123 UCUACUGCCAACAAUAUAGAGAUA-UCCCUCUGUG-------UGUGCAAUUGCAAAUUGCGCAU-UUUUGUUGCAUUUAC--CAUAGUUUUCGUUGUUGCCGAUUGUGUUGGCGGCAGGCCAAAUGAUUUGGCAAGGAAA (((.(((((((((((((((((...-...))))))(-------((((((((.....)))))))))-...............--..........))))))((((((...)))))))))))(((((.....)))))..))).. ( -42.00, z-score = -2.47, R) >droYak2.chrX 8573232 139 - 21770863 UUCACUGCCAACAAUAUAGGGAUAUUCACCCUGUGUUUUCUCUGUGCAAUUGCAAAUUGCGCAU-UUUUGUUGCAUUUGUUGCAUCGUUUUCGUUGUUGCCGAUUGUGUUGGCGGCAGGCCAAAUGAUUUGGCAAGGAAA (((..((((((.(((((((((.......))))))))).....((((((((.....)))))))).-......((((.....))))((((((...(((((((((((...)))))))))))...)))))).))))))..))). ( -46.30, z-score = -2.27, R) >droEre2.scaffold_4690 6131537 125 + 18748788 -----UGCCAACACUAUACAUA---CUUGUAUAUACCCUUACUGUG---UUGCAAG-UGCGCAU-UUUUGUUGCAUUCAG--CCUGGGAUUUGUUGUUGCCGAUUGUGUUGGCGGCAGGCCAAAUGAUUUGGCAAGGAAA -----((((((...((((((..---..))))))..(((...(((((---(.(((((-.......-.))))).)))..)))--...)))((((((((((((((((...)))))))))))..)))))...))))))...... ( -37.30, z-score = -0.41, R) >droAna3.scaffold_13248 2038818 116 - 4840945 --------CAUAAAGAGAGAGAUA-----UAUAUA--------AUUUCGUUGCAGAUUGCAGAUGCUUUGUGGCAACAGACUGGGGACUGGGAGUGAGGCAG---GGACUGGCGGCAGGCCAAGUUAUUUGGCAAGAAGC --------..........(((((.-----......--------))))).((((.....))))..(((((.((.(((..((((..((.(((...((.((....---...)).))..))).)).))))..))).)).))))) ( -26.60, z-score = -0.29, R) >consensus U__ACUGCCAACAAUAUAGAGAUA_UCCCUCUGUG___U__CUGUGCAAUUGCAAAUUGCGCAU_UUUUGUUGCAUUUAC__CAUCGUUUUCGUUGUUGCCGAUUGUGUUGGCGGCAGGCCAAAUGAUUUGGCAAGGAAA ........(((((((((((((.......))))))).......(((((((((...)))))))))....))))))....................((((((((((.....))))))))))((((((...))))))....... (-22.77 = -25.14 + 2.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:19 2011