
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,326,301 – 14,326,492 |
| Length | 191 |
| Max. P | 0.951665 |

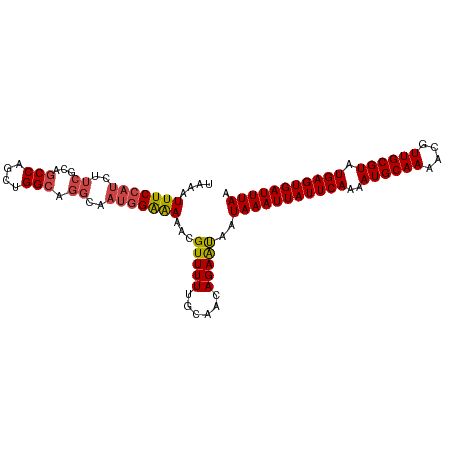
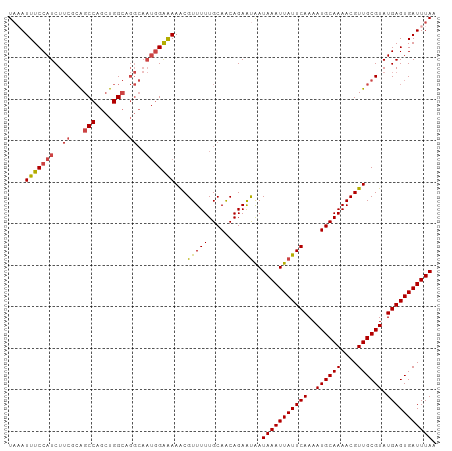
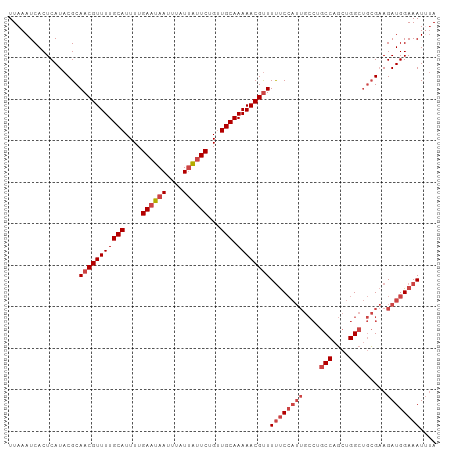
| Location | 14,326,301 – 14,326,404 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.97 |
| Shannon entropy | 0.15561 |
| G+C content | 0.36141 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.35 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14326301 103 + 22422827 UAAAUUUCCAUCUUCGCAGCCAGCUGGCUGGCAAUGGAAAAACGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAA ((((((.((((...(((.(((((....)))))........((((((((.(((...((((((....))))))....)))))))))))))).))).).)))))). ( -29.80, z-score = -3.42, R) >droSim1.chrX 11017844 103 + 17042790 UAAAUUUCCAUCUUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAA ....(((((((..((...(((....))).))..))))))).....................(((((((((((..((((((....)))))).))))))))))). ( -25.70, z-score = -2.14, R) >droSec1.super_20 921545 103 + 1148123 UAAAUUUCCAUCUUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAACAGAGCAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAA ....(((((((..((...(((....))).))..))))))).......((((......))))(((((((((((..((((((....)))))).))))))))))). ( -28.50, z-score = -2.63, R) >droYak2.chrX 8563602 103 + 21770863 UAAAUUUCCAUCUUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAGCAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAA ((((((.((((...(((.(((....)))............((((((((.(((...((((((....))))))....)))))))))))))).))).).)))))). ( -26.10, z-score = -1.84, R) >droEre2.scaffold_4690 6122564 103 - 18748788 UACAUUUCCAUCUUCGCAGCCAGCUGGCUGGCAAUGGAAAAGCGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAA ....(((((((...(.(((((....))))))..))))))).(((....)))..........(((((((((((..((((((....)))))).))))))))))). ( -30.70, z-score = -3.19, R) >droAna3.scaffold_13248 2027371 94 + 4840945 UCCAUUUCCAUCUACCAGGCCAGAUGG---------GGGAAACGUUUUUGCAACAGAAUGAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAA ....(..((((((........))))))---------..)...((((((......)))))).(((((((((((..((((((....)))))).))))))))))). ( -25.30, z-score = -2.48, R) >consensus UAAAUUUCCAUCUUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAA ....(((((((..((...(((....))).))..)))))))...(((((......)))))..(((((((((((..((((((....)))))).))))))))))). (-21.90 = -22.35 + 0.45)

| Location | 14,326,301 – 14,326,404 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 91.97 |
| Shannon entropy | 0.15561 |
| G+C content | 0.36141 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -20.00 |
| Energy contribution | -21.53 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14326301 103 - 22422827 UUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGUUGCAAAAACGUUUUUCCAUUGCCAGCCAGCUGGCUGCGAAGAUGGAAAUUUA .................(((((((((((....((((((....))))))...))).))))))))((((((((..(((((....)))))....)))))))).... ( -30.20, z-score = -3.93, R) >droSim1.chrX 11017844 103 - 17042790 UUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGUUGCAAAAACGUUUUUCCAUUGCCUGCCAGCUGGCUGCGAAGAUGGAAAUUUA .................(((((((((((....((((((....))))))...))).))))))))((((((((((..(((....))).))...)))))))).... ( -27.00, z-score = -2.93, R) >droSec1.super_20 921545 103 - 1148123 UUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUGCUCUGUUGCAAAAACGUUUUUCCAUUGCCUGCCAGCUGGCUGCGAAGAUGGAAAUUUA .................(((((((((((....((.(((....))).))...))).))))))))((((((((((..(((....))).))...)))))))).... ( -23.00, z-score = -1.12, R) >droYak2.chrX 8563602 103 - 21770863 UUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGCUGCAAAAACGUUUUUCCAUUGCCUGCCAGCUGGCUGCGAAGAUGGAAAUUUA .................(((((((((((....((((((....))))))...))).))))))))((((((((((..(((....))).))...)))))))).... ( -26.30, z-score = -2.54, R) >droEre2.scaffold_4690 6122564 103 + 18748788 UUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGUUGCAAAAACGCUUUUCCAUUGCCAGCCAGCUGGCUGCGAAGAUGGAAAUGUA ...............(((.(((((((((....((((((....))))))...))).))))))..((((((((..(((((....)))))....))))))))))). ( -29.20, z-score = -3.14, R) >droAna3.scaffold_13248 2027371 94 - 4840945 UUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUCAUUCUGUUGCAAAAACGUUUCCC---------CCAUCUGGCCUGGUAGAUGGAAAUGGA .........((((..(.(((((((((((....((((........))))...))).)))))))).)..---------(((((((......)))))))..)))). ( -20.60, z-score = -1.37, R) >consensus UUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGUUGCAAAAACGUUUUUCCAUUGCCUGCCAGCUGGCUGCGAAGAUGGAAAUUUA .................(((((((((((....((((((....))))))...))).))))))))((((((((....(((....)))......)))))))).... (-20.00 = -21.53 + 1.53)
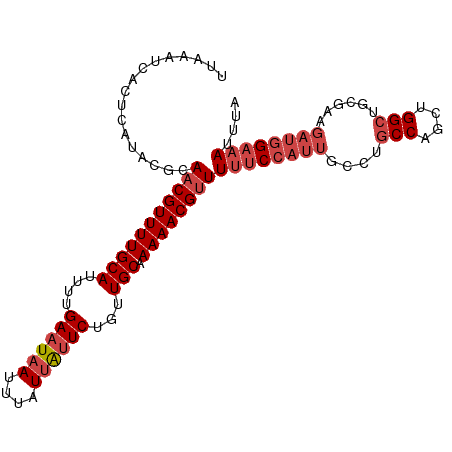
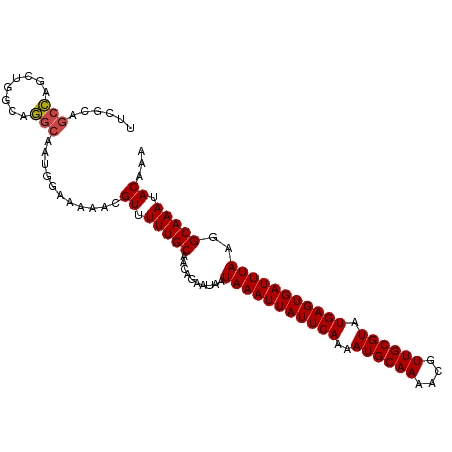
| Location | 14,326,313 – 14,326,416 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.33 |
| Shannon entropy | 0.16823 |
| G+C content | 0.36434 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.22 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14326313 103 + 22422827 UUCGCAGCCAGCUGGCUGGCAAUGGAAAAACGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACAAA ((((..(((((....)))))..)))).....((.(((((..........(((((((((((..((((((....)))))).)))))))))))..))))).))... ( -28.10, z-score = -2.58, R) >droSim1.chrX 11017856 103 + 17042790 UUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACAAA ......(((..((((((((.((((......)))))))))..))).....(((((((((((..((((((....)))))).))))))))))).)))......... ( -27.00, z-score = -2.48, R) >droSec1.super_20 921557 103 + 1148123 UUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAACAGAGCAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACAAA ......(((.(((.(((((.((((......))))))))).....)))..(((((((((((..((((((....)))))).))))))))))).)))......... ( -28.10, z-score = -2.42, R) >droYak2.chrX 8563614 103 + 21770863 UUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAGCAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACAAA ......(((.((((.((((.((((......)))))))))))).......(((((((((((..((((((....)))))).))))))))))).)))......... ( -30.00, z-score = -3.23, R) >droEre2.scaffold_4690 6122576 103 - 18748788 UUCGCAGCCAGCUGGCUGGCAAUGGAAAAGCGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAAGCAAAUACACA ..(((.(((((....)))))........((((((((.(((...((((((....))))))....)))))))))))))).....(((((((.....)))).))). ( -28.20, z-score = -2.26, R) >droAna3.scaffold_13248 2027383 94 + 4840945 --------UACCAGGCCAG-AUGGGGGAAACGUUUUUGCAACAGAAUGAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAAGCAAAUACAAA --------..(((......-.))).(....)((.(((((..(.....).(((((((((((..((((((....)))))).)))))))))))..))))).))... ( -22.60, z-score = -2.70, R) >consensus UUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACAAA ......(((........)))...........((.(((((..........(((((((((((..((((((....)))))).)))))))))))..))))).))... (-20.28 = -20.22 + -0.07)

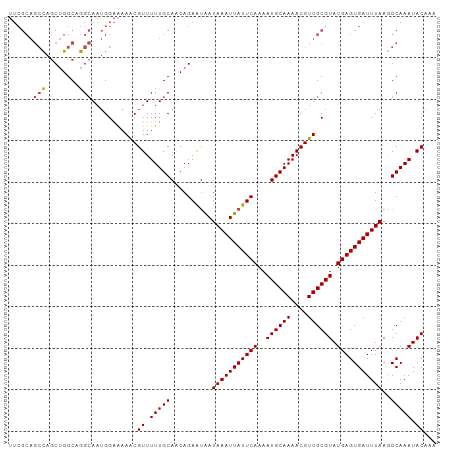
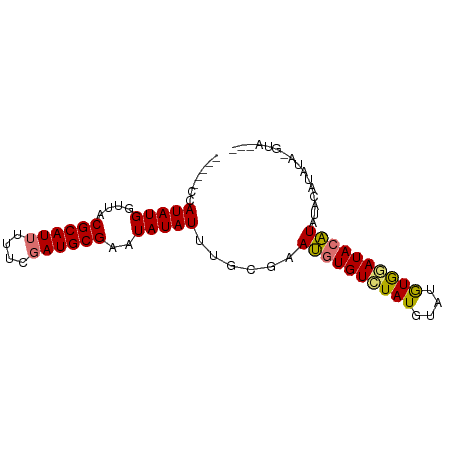
| Location | 14,326,416 – 14,326,492 |
|---|---|
| Length | 76 |
| Sequences | 4 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Shannon entropy | 0.37313 |
| G+C content | 0.34388 |
| Mean single sequence MFE | -18.07 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.32 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14326416 76 - 22422827 -----ACAUAUGGUUACGCAUUUUUCGAUGCGAAUAUAUUUGCGAAUGUGUCUAUGUAAGUGGAUACAUAUACAUAUA-GUA--- -----(((((((....((((((....)))))).............((((((((((....))))))))))...))))).-)).--- ( -20.30, z-score = -2.02, R) >droSim1.chrX 11017959 76 - 17042790 -----CCAUAUGGUUACGCAUUUUUCGAUGCGAAUAUAUUUGCGAAUGUGUCUAUGUAUGUGGAUACAUAUACAUAUA-GUA--- -----..(((((....((((((....)))))).............((((((((((....))))))))))...))))).-...--- ( -20.00, z-score = -1.50, R) >droYak2.chrX 8563717 80 - 21770863 -----CCAUAUGGUUACGCAUUUUUCCAUGCGAAUAUAUUUGCGAAUGUGUCUAUGCAUGUACAUACAUACAUACAUAUAUAGUA -----..(((((....(((((......))))).............((((((.((((......)))).)))))).)))))...... ( -15.40, z-score = 0.10, R) >droEre2.scaffold_4690 6122679 69 + 18748788 CCAUCCCAUAUGGGCACGCAUUUUCCGAUGCGAAUAUAUUUGCGAACGUGUAUAUGUACAUGUAUA-GUA--------------- ....(((....)))..((((((....))))))........(((..(((((((....)))))))...-)))--------------- ( -16.60, z-score = -0.61, R) >consensus _____CCAUAUGGUUACGCAUUUUUCGAUGCGAAUAUAUUUGCGAAUGUGUCUAUGUAUGUGGAUACAUAUACAUAUA_GUA___ .......(((((....((((((....))))))..)))))......((((((((((....))))))))))................ (-14.57 = -14.32 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:15 2011