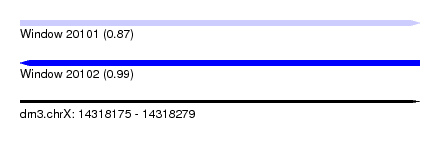
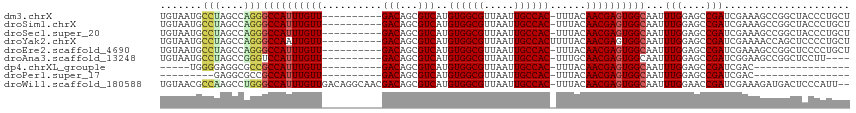
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,318,175 – 14,318,279 |
| Length | 104 |
| Max. P | 0.988259 |

| Location | 14,318,175 – 14,318,279 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.67 |
| Shannon entropy | 0.33750 |
| G+C content | 0.52069 |
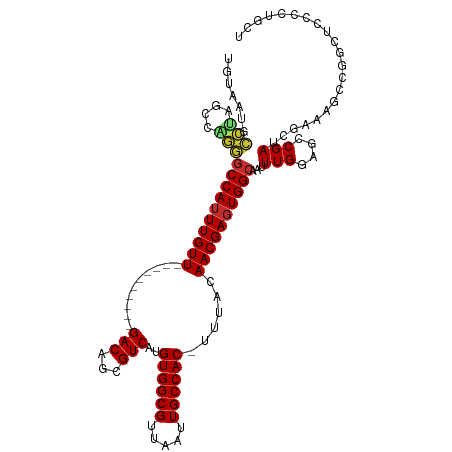
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -18.21 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
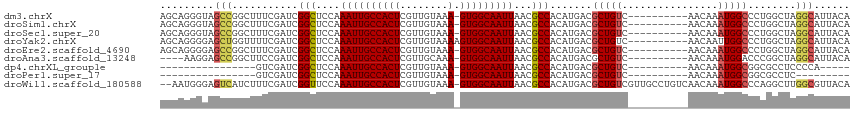
>dm3.chrX 14318175 104 + 22422827 AGCAGGGUAGCCGGCUUUCGAUCGGCUCCAAAUUGCCACUCGUUGUAAA-GUGGCAAUUAACGCCACAUGACGCUGUC----------AACAAAUGGCCCUGGCUAGGCAUUACA .....((.(((((((....).)))))))).((((((((((........)-)))))))))...(((...((((...)))----------).....((((....)))))))...... ( -36.00, z-score = -1.66, R) >droSim1.chrX 11009449 104 + 17042790 AGCAGGGUAGCCGGCUUUCGAUCGGCUCCAAAUUGCCACUCGUUGUAAA-GUGGCAAUUAACGCCACAUGACGCUGUC----------AACAAAUGGCCCUGGCUAGGCAUUACA .....((.(((((((....).)))))))).((((((((((........)-)))))))))...(((...((((...)))----------).....((((....)))))))...... ( -36.00, z-score = -1.66, R) >droSec1.super_20 913581 104 + 1148123 AGCAGGGUAGCCGGCUUUCGAUCGGCUCCAAAUUGCCACUCGUUGUAAA-GUGGCAAUUAACGCCACAUGACGCUGUC----------AACAAAUGGCCCUGGCUAGGCAUUACA .....((.(((((((....).)))))))).((((((((((........)-)))))))))...(((...((((...)))----------).....((((....)))))))...... ( -36.00, z-score = -1.66, R) >droYak2.chrX 8555994 105 + 21770863 AGCAGGGGAGCUGGUUUUCGAUCGGCUCCAAAUUGCCACUCGUUGUAAAAGUGGCAAUUAACGCCACAUGACGCUGUC----------AACAAUUGGCCCUGGCUAGGCAUUACA ((((((((((((((((...)))))))))).....((((...(((((....(((((.......))))).((((...)))----------))))))))))))).))).......... ( -38.80, z-score = -2.58, R) >droEre2.scaffold_4690 6115119 104 - 18748788 AGCAGGGGAGCCGGCUUUCGAUCGGCUCCAAAUUGCCACUCGUUGUAAA-GUGGCAAUUAACGCCACAUGACGCUGUC----------AACAAAUGGCCCUGGCUAGGCAUUACA ......(((((((((....).)))))))).((((((((((........)-)))))))))...(((...((((...)))----------).....((((....)))))))...... ( -39.80, z-score = -2.60, R) >droAna3.scaffold_13248 2020226 100 + 4840945 ----AAGGAGCCGGCUUCCGAUCGGCUCCAAAUUGCCACUCGUUGCAAA-GUGGCAAUUAACGCCACAUGACGCUGUC----------AACAAAUGGACCCGGCUAGGCAUUACA ----..(((((((((....).)))))))).((((((((((........)-)))))))))...(((...((((...)))----------).....(((......))))))...... ( -35.20, z-score = -2.43, R) >dp4.chrXL_group1e 7566490 83 - 12523060 ----------------GUCGAUCGGCUCCAAAUUGCCACUCGUUGUAAA-GUGGCAAUUAACGCCACAUGACGCUGUC----------AACAAAUGGCGGCGCCUCCCCA----- ----------------((((...(((....((((((((((........)-)))))))))...)))...))))((((((----------(.....))))))).........----- ( -30.70, z-score = -3.49, R) >droPer1.super_17 15627 79 - 1930428 ----------------GUCGAUCGGCUCCAAAUUGCCACUCGUUGUAAA-GUGGCAAUUAACGCCACAUGACGCUGUC----------AACAAAUGGCGGCGCCUC--------- ----------------((((...(((....((((((((((........)-)))))))))...)))...))))((((((----------(.....))))))).....--------- ( -30.70, z-score = -3.89, R) >droWil1.scaffold_180588 109185 112 + 1294757 --AAUGGGAGUCAUCUUUCGAUCGGUUCCAAAUUGCCACUCGUUGUAAA-GUGGCAAUUAACGCCACAUGACGCUGUCGUUGCCUGUCAACAAAUGGCCCAGGCUUGGCGUUACA --....(((..((((....))).)..))).((((((((((........)-)))))))))(((((((.(((((...))))).((((((((.....))))..)))).)))))))... ( -39.70, z-score = -2.81, R) >consensus AGCAGGGGAGCCGGCUUUCGAUCGGCUCCAAAUUGCCACUCGUUGUAAA_GUGGCAAUUAACGCCACAUGACGCUGUC__________AACAAAUGGCCCUGGCUAGGCAUUACA .........(((...........(((....(((((((((...........)))))))))...))).......(((((................)))))........)))...... (-18.21 = -18.60 + 0.39)
| Location | 14,318,175 – 14,318,279 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
| Shannon entropy | 0.33750 |
| G+C content | 0.52069 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.21 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
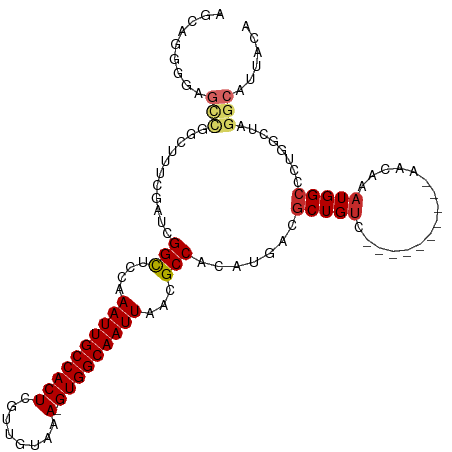
>dm3.chrX 14318175 104 - 22422827 UGUAAUGCCUAGCCAGGGCCAUUUGUU----------GACAGCGUCAUGUGGCGUUAAUUGCCAC-UUUACAACGAGUGGCAAUUUGGAGCCGAUCGAAAGCCGGCUACCCUGCU ..........(((.(((((((......----------...((((((....))))))(((((((((-((......))))))))))))))(((((..(....).))))).))))))) ( -40.20, z-score = -2.84, R) >droSim1.chrX 11009449 104 - 17042790 UGUAAUGCCUAGCCAGGGCCAUUUGUU----------GACAGCGUCAUGUGGCGUUAAUUGCCAC-UUUACAACGAGUGGCAAUUUGGAGCCGAUCGAAAGCCGGCUACCCUGCU ..........(((.(((((((......----------...((((((....))))))(((((((((-((......))))))))))))))(((((..(....).))))).))))))) ( -40.20, z-score = -2.84, R) >droSec1.super_20 913581 104 - 1148123 UGUAAUGCCUAGCCAGGGCCAUUUGUU----------GACAGCGUCAUGUGGCGUUAAUUGCCAC-UUUACAACGAGUGGCAAUUUGGAGCCGAUCGAAAGCCGGCUACCCUGCU ..........(((.(((((((......----------...((((((....))))))(((((((((-((......))))))))))))))(((((..(....).))))).))))))) ( -40.20, z-score = -2.84, R) >droYak2.chrX 8555994 105 - 21770863 UGUAAUGCCUAGCCAGGGCCAAUUGUU----------GACAGCGUCAUGUGGCGUUAAUUGCCACUUUUACAACGAGUGGCAAUUUGGAGCCGAUCGAAAACCAGCUCCCCUGCU ..........(((.(((((((.....(----------(((...))))..))))...(((((((((((.......))))))))))).(((((.(.........).))))))))))) ( -33.80, z-score = -1.65, R) >droEre2.scaffold_4690 6115119 104 + 18748788 UGUAAUGCCUAGCCAGGGCCAUUUGUU----------GACAGCGUCAUGUGGCGUUAAUUGCCAC-UUUACAACGAGUGGCAAUUUGGAGCCGAUCGAAAGCCGGCUCCCCUGCU ..........(((.(((((((((((((----------(((...)))..((((((.....))))))-.....)))))))))).....(((((((..(....).))))))))))))) ( -44.90, z-score = -4.16, R) >droAna3.scaffold_13248 2020226 100 - 4840945 UGUAAUGCCUAGCCGGGUCCAUUUGUU----------GACAGCGUCAUGUGGCGUUAAUUGCCAC-UUUGCAACGAGUGGCAAUUUGGAGCCGAUCGGAAGCCGGCUCCUU---- ..(((((((((..((.(((........----------)))..))...)).)))))))((((((((-((......))))))))))..(((((((..(....).)))))))..---- ( -39.60, z-score = -2.55, R) >dp4.chrXL_group1e 7566490 83 + 12523060 -----UGGGGAGGCGCCGCCAUUUGUU----------GACAGCGUCAUGUGGCGUUAAUUGCCAC-UUUACAACGAGUGGCAAUUUGGAGCCGAUCGAC---------------- -----..((..(((.(((((((....(----------(((...)))).)))))...(((((((((-((......))))))))))).)).)))..))...---------------- ( -34.40, z-score = -3.30, R) >droPer1.super_17 15627 79 + 1930428 ---------GAGGCGCCGCCAUUUGUU----------GACAGCGUCAUGUGGCGUUAAUUGCCAC-UUUACAACGAGUGGCAAUUUGGAGCCGAUCGAC---------------- ---------..(((.(((((((....(----------(((...)))).)))))...(((((((((-((......))))))))))).)).))).......---------------- ( -32.90, z-score = -3.75, R) >droWil1.scaffold_180588 109185 112 - 1294757 UGUAACGCCAAGCCUGGGCCAUUUGUUGACAGGCAACGACAGCGUCAUGUGGCGUUAAUUGCCAC-UUUACAACGAGUGGCAAUUUGGAACCGAUCGAAAGAUGACUCCCAUU-- ..((((((((.(((((...((.....)).)))))...(((...)))...))))))))((((((((-((......))))))))))..(((..(.(((....))))..)))....-- ( -41.00, z-score = -3.55, R) >consensus UGUAAUGCCUAGCCAGGGCCAUUUGUU__________GACAGCGUCAUGUGGCGUUAAUUGCCAC_UUUACAACGAGUGGCAAUUUGGAGCCGAUCGAAAGCCGGCUCCCCUGCU .......(((....)))((((((((((..........(((...)))..((((((.....))))))......))))))))))...(((....)))..................... (-22.10 = -22.21 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:12 2011