| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,306,890 – 14,306,982 |
| Length | 92 |
| Max. P | 0.965746 |

| Location | 14,306,890 – 14,306,982 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.13 |
| Shannon entropy | 0.55590 |
| G+C content | 0.48652 |
| Mean single sequence MFE | -12.53 |
| Consensus MFE | -8.57 |
| Energy contribution | -8.61 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
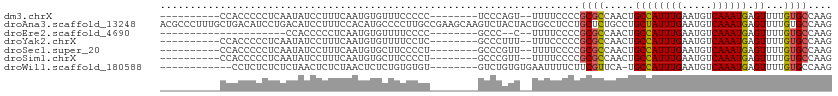
>dm3.chrX 14306890 92 - 22422827 ----------CCACCCCCUCAAUAUCCUUUCAAUGUGUUUCCCCC--------UCCCAGU--UUUUCCCCGCGCCAACUGCCAUUUGAAUGUCAAAUGAGUUUUGUGCCAAG ----------...................................--------.......--........((((.((((..((((((.....))))))))))..)))).... ( -10.90, z-score = -1.53, R) >droAna3.scaffold_13248 2012601 112 - 4840945 ACGCCCUUUGCUGACAUCCUGACAUCCUUUCCACAUGCCCCUUGCCGAAGCAAGUCUACUACUGCCUCCUGCUCUGCCUGCUAUUUGAAUGUCAAAUGAGUUUUGUGCCAAG ..((.(...(((..(((..((((((.(......((.((..(((((....))))).........((.....))...)).))......).)))))).))))))...).)).... ( -16.00, z-score = 0.45, R) >droEre2.scaffold_4690 6108849 79 + 18748788 ---------------------CCACCCCCUCAAUGUGUUUUCCCC--------GCCC--C--UUUUCCCCGCGCCAACUGCCAUUUGAAUGUCAAAUGAGUUUUGUGCCAAG ---------------------........................--------....--.--........((((.((((..((((((.....))))))))))..)))).... ( -10.90, z-score = -1.44, R) >droYak2.chrX 8549330 92 - 21770863 ----------CCACCCCCUCAAUAUCCUUUCAAUGUGUUUUCCUC--------GCCCUUU--UUUCCCCCGCGCCAACUGCCAUUUGAAUGUCAAAUGAGUUUUGUGCCAAG ----------........................(((.......)--------)).....--........((((.((((..((((((.....))))))))))..)))).... ( -11.20, z-score = -1.58, R) >droSec1.super_20 907423 92 - 1148123 ----------CCACCCCCUCAAUAUCCUUUCAAUGUGCUUCCCCU--------GCCCGUU--UUUUCCCCGCGCCAACUGCCAUUUGAAUGUCAAAUGAGUUUUGUGCCAAG ----------.....................((((.((.......--------)).))))--........((((.((((..((((((.....))))))))))..)))).... ( -13.50, z-score = -1.93, R) >droSim1.chrX 11002516 92 - 17042790 ----------CCACCCCCUCAAUAUCCUUUCAAUGUGCUUCCCCU--------GCCCGUU--UUUUCCCCGCGCCAACUGCCAUUUGAAUGUCAAAUGAGUUUUGUGCCAAG ----------.....................((((.((.......--------)).))))--........((((.((((..((((((.....))))))))))..)))).... ( -13.50, z-score = -1.93, R) >droWil1.scaffold_180588 1189055 91 + 1294757 ------------CCUCUCUCUCUAACUCUCUAACUCUCUGUGUGU--------GUCUGUGUGAAUUUUCUUCGUUCA-UGCCAUUUGAAUGUCAAAUGAGUUUUGUGCCAAG ------------..........................((.(..(--------(.((((((((((.......)))))-)))((((((.....))))))))...))..))).. ( -11.70, z-score = -0.86, R) >consensus __________CCACCCCCUCAAUAUCCUUUCAAUGUGCUUCCCCC________GCCCGUU__UUUUCCCCGCGCCAACUGCCAUUUGAAUGUCAAAUGAGUUUUGUGCCAAG ......................................................................((((.....((((((((.....)))))).))...)))).... ( -8.57 = -8.61 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:09 2011