
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,298,287 – 14,298,396 |
| Length | 109 |
| Max. P | 0.657545 |

| Location | 14,298,287 – 14,298,396 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.12 |
| Shannon entropy | 0.52692 |
| G+C content | 0.54022 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.23 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14298287 109 + 22422827 CACACACAAAAUUAAAAGCCAAUUAGGCGGUGACGAGAGCCCAAAGGUUGC--CCCUUUUUCCCCAUAACUCACGCGGUUGCCC------CACCCGCGUUGAAUGCAACCGCUCCUG .................(((.....)))(....)..((((.....((((((--.................((((((((.((...------)).))))).)))..))))))))))... ( -25.91, z-score = -1.00, R) >droAna3.scaffold_13248 2002069 106 + 4840945 --CGCGCAAAAUUAAAAGCCAAUUAAGCGUUGAAGAGUCUCACGAGUCCUGAGUCGUCCUGGCCUCCAA---AUGCAGCCAGGU------CCUUGGUGGGGCUCCUGCUCGCUCUUG --.((((..((((.......))))..))))..((((((..((.((((((((....(.((((((......---.....)))))).------).....)))))))).))...)))))). ( -32.50, z-score = -0.82, R) >droEre2.scaffold_4690 6101478 96 - 18748788 CACACACAAAAUUAAAAGCCAAUUAGGCGGUGACGAGAGCCCGAGGG---------------CCCAUAACUCAAGCGGUUGCCC------CACCCGCGUGAAAUGCAACCGCUCCUG .................(((.....)))(....)(((.(((....))---------------)......))).(((((((((..------(((....)))....))))))))).... ( -28.40, z-score = -1.80, R) >droYak2.chrX 8540886 108 + 21770863 CACACACAAAAUUAAAAGCCAAUUAGGCGGUGACGAGAAGGAGGCGG---------CUUUUUCCCAUAACUCACGCGGUUUCUCUCCAUUCCCCCGCAUGAAAUGCAACCGCUCCUG .................(((.....)))(....)....(((((.(((---------........(((...(((.((((...............)))).))).)))...)))))))). ( -22.56, z-score = 0.35, R) >droSec1.super_20 898988 111 + 1148123 CACACACAAAAUUAAAAGCCAAUUAGGCGGUGACGAGAGCCCCAAGUAUGAUGCCCCUUUUUCCCAUAACUCACGCGGUUGCCC------CACCCGCGUGGAAUGCAACCGCUCCUG .................(((.....)))((..(.(((.((..((....))..))..))).)..)).........((((((((.(------(((....))))...))))))))..... ( -29.20, z-score = -1.99, R) >droSim1.chrX 10992766 111 + 17042790 CACACACAAAAUUAAAAGCCAAUUAGGCGGUGACGAGAGCCCCAAGUAUGAUGCCCCUUUUUCCCAUAACUCACGCGGUUGCCC------CACCCGCGUGGAAUGCAACCGCUCCUG .................(((.....)))((..(.(((.((..((....))..))..))).)..)).........((((((((.(------(((....))))...))))))))..... ( -29.20, z-score = -1.99, R) >consensus CACACACAAAAUUAAAAGCCAAUUAGGCGGUGACGAGAGCCCCAAGG_UG___CCCCUUUUUCCCAUAACUCACGCGGUUGCCC______CACCCGCGUGGAAUGCAACCGCUCCUG .................(((.....)))..............................................((((((((......................))))))))..... (-14.48 = -14.23 + -0.25)

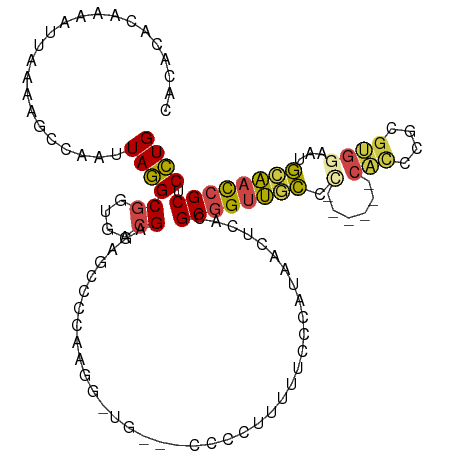
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:08 2011