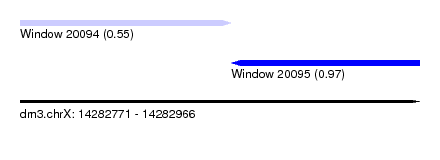
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,282,771 – 14,282,966 |
| Length | 195 |
| Max. P | 0.966001 |

| Location | 14,282,771 – 14,282,874 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.29 |
| Shannon entropy | 0.12238 |
| G+C content | 0.53552 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -31.24 |
| Energy contribution | -31.16 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.550086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
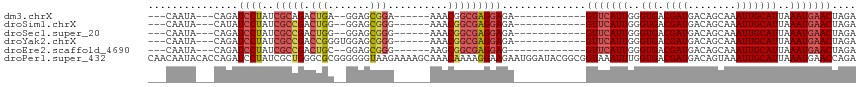
>dm3.chrX 14282771 103 + 22422827 UCCCAGGAGCAUCGCAGUGAGCACGGAGCCUCUUCUGCUCACUUACUGUUUUACAUUUGAAUGGCUCGGCAGGUCUCCAAUCAGUCUCGCCUGGAGGCAUUCU .....((((((....((((((((.((((...))))))))))))...))))))......(((((.((((((..(.((......)).)..)))..))).))))). ( -34.10, z-score = -1.22, R) >droSim1.chrX 10976317 98 + 17042790 UCCCAGGAGCAUCGCAGUGAGCA-GGAGC-UCUUCUGCUCACUUACUGUUU-ACAUU-GGAUGGCUCGGCAGGUCUCCAAUCAGUCUCGCCUGGAGGCAUUU- ..(((((........((((((((-(((..-..))))))))))).((((...-..(((-(((.((((.....))))))))))))))....)))))........- ( -36.20, z-score = -1.62, R) >droSec1.super_20 883613 102 + 1148123 UCCCAGGAGCAUCGCAGUGAGCGGGAGGC-UCUCCUGCUCACUUACUGUUUUACAUUUGAAUGGCUCGGCAGGUCUCCAAUCAGUCUCGCCUGGAGGCAUUCU .....((((((....((((((((((((..-.))))))))))))...))))))......(((((.((((((..(.((......)).)..)))..))).))))). ( -40.10, z-score = -2.29, R) >droYak2.chrX 8523899 102 + 21770863 UCCCAGGAGCAUCGCAGCGAGCACGAGGCU-CUUCUGCUCACUUACUGUUUUACAUUUGAAUGGCUCGGCAGGUCUCCAAUCAGUCUCGCCUGGAGGCAUUCU .....((((((....((.(((((.(((...-.)))))))).))...))))))......(((((.((((((..(.((......)).)..)))..))).))))). ( -29.50, z-score = 0.07, R) >droEre2.scaffold_4690 6085600 103 - 18748788 UCCCAGGAGCAUCGCAGUGAGCAAGAGCCUUCUUCUGCUCACUUACUGUUUUACAUUUGAAUGGCUCGGCAGGUCUCCAAUCAGUCUCGCCUGGAGGCAUUCU .....((((((....((((((((..((....))..))))))))...))))))......(((((.((((((..(.((......)).)..)))..))).))))). ( -33.80, z-score = -1.40, R) >consensus UCCCAGGAGCAUCGCAGUGAGCACGAGGC_UCUUCUGCUCACUUACUGUUUUACAUUUGAAUGGCUCGGCAGGUCUCCAAUCAGUCUCGCCUGGAGGCAUUCU .....((((((....((((((((.((((...))))))))))))...))))))......(((((.((((((..(.((......)).)..)))..))).))))). (-31.24 = -31.16 + -0.08)
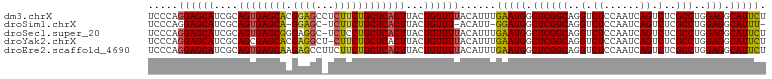
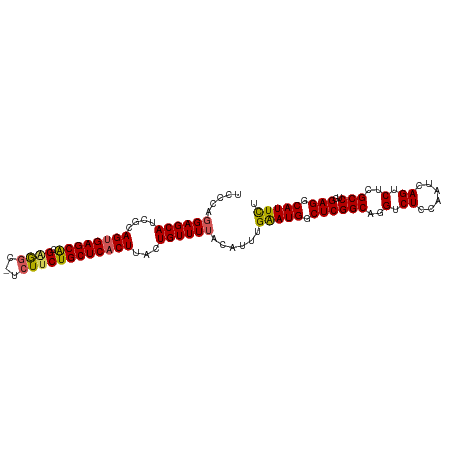
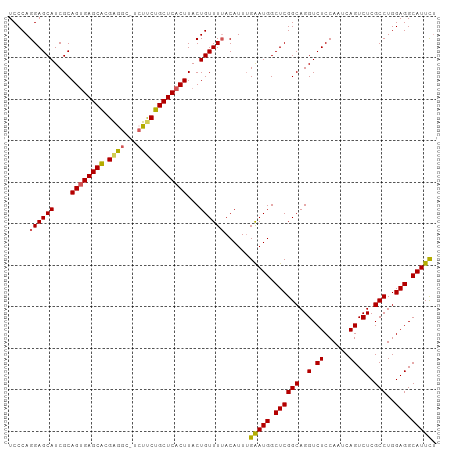
| Location | 14,282,874 – 14,282,966 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.21 |
| Shannon entropy | 0.29703 |
| G+C content | 0.47835 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
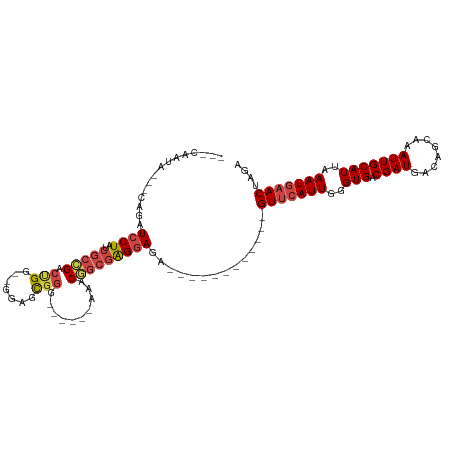
>dm3.chrX 14282874 92 - 22422827 ---CAAUA---CAGAUCCUAUCGCAGACUGA--GGAGCGGA------AAACGGCGAGGAGA------------GUUCAUUGGGUGACGAUGACAGCAAAUUGCAUUAAAUGAACUAGA ---.....---....(((..((((...(((.--....))).------.....))))))).(------------(((((((..(((.((((........)))))))..))))))))... ( -21.90, z-score = -1.84, R) >droSim1.chrX 10976415 92 - 17042790 ---CAAUA---CAUAUCCUAUCGCCGACUGG--GGAGCGGG------AAACGGCGAGGAGA------------GUUCAUUGGGUGACGAUGACAGCAAAUUGCAUUAAAUGAACUAGA ---.....---....(((..((((((.((.(--....).))------...))))))))).(------------(((((((..(((.((((........)))))))..))))))))... ( -28.90, z-score = -3.53, R) >droSec1.super_20 883715 92 - 1148123 ---CAAUA---CAGAUCCUAUCGCCGACUGG--GGAGCGGG------AAACGGCGAGGAGA------------GUUCAUUGGGUGACGAUGACAGCAAAUUGCAUUAAAUGAACUAGA ---.....---....(((..((((((.((.(--....).))------...))))))))).(------------(((((((..(((.((((........)))))))..))))))))... ( -28.90, z-score = -3.43, R) >droYak2.chrX 8524001 94 - 21770863 ---CAAUA---CAGAUCCUAUCGCCGACCGGGUGGAGCGGG------AAACGGCGAGGAGA------------GUUCAUUGGGUGACGAUGACAGCAAAUUGCAUUAAAUGAACUAGA ---.....---....((((.(((((.....))))).((.(.------...).)).)))).(------------(((((((..(((.((((........)))))))..))))))))... ( -30.50, z-score = -3.20, R) >droEre2.scaffold_4690 6085703 91 + 18748788 ---CAAUA---CAGAUCCUAUCGCCGACUGC--GGAGCGGG------AAGCGGCGAGGAG-------------GUUCAUUGGGUGACGAUGACAGCAAAUUGCAUUAAAUGAACUAGA ---.....---....(((..((((((.((((--...)))).------...)))))))))(-------------(((((((..(((.((((........)))))))..))))))))... ( -31.20, z-score = -3.46, R) >droPer1.super_432 18861 118 + 20690 CAACAAUACACCAGAUCCUAUCGCUGGGCGCGGGGGGUAAGAAAAGCAAACAAAAGGAAGAAUGGAUACGGCGGUAAAUUUGGUGACGAUGACAGUAAAUUGCAUUAAAUGAACCAGA ..........(((..((((.((((.....))))...((.......)).......))))....))).......(((..((((((((.((((........))))))))))))..)))... ( -22.80, z-score = -0.12, R) >consensus ___CAAUA___CAGAUCCUAUCGCCGACUGG__GGAGCGGG______AAACGGCGAGGAGA____________GUUCAUUGGGUGACGAUGACAGCAAAUUGCAUUAAAUGAACUAGA ...............((((..(((((.(((.......)))..........)))))))))..............(((((((..(((.((((........)))))))..))))))).... (-18.08 = -18.97 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:06 2011