
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,275,180 – 14,275,325 |
| Length | 145 |
| Max. P | 0.592563 |

| Location | 14,275,180 – 14,275,325 |
|---|---|
| Length | 145 |
| Sequences | 7 |
| Columns | 187 |
| Reading direction | reverse |
| Mean pairwise identity | 55.58 |
| Shannon entropy | 0.77275 |
| G+C content | 0.50084 |
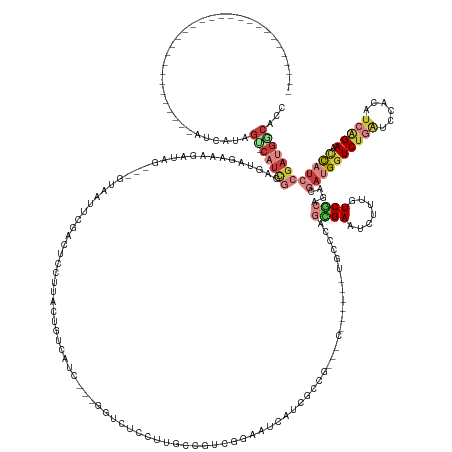
| Mean single sequence MFE | -46.46 |
| Consensus MFE | -7.01 |
| Energy contribution | -7.49 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.70 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.15 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.592563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
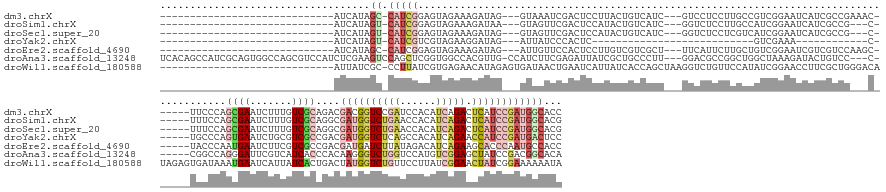
>dm3.chrX 14275180 145 - 22422827 -----------------------------AUCAUAGC-CAUCGGAGUAGAAAGAUAG---GUAAAUCGACUCCUUACUGUCAUC---GUCCUCCUUGCCGUCGGAAUCAUCGCCGAAAC------UUCCCAGCGAAUCUUUGUCGCAGACGACGGUCCGAUCCACAUCAGACUCAUCCGAUGGCACC -----------------------------......((-(((((((..((...(((.(---((((...(((........)))...---(....).)))))((((((.((.((((.(....------....).))))......((((....))))))))))))....)))...))..)))))))))... ( -41.80, z-score = -1.46, R) >droSim1.chrX 10968405 142 - 17042790 -----------------------------AUCAUAGU-CAUCGGAGUAGAAAGAUAA---GUAGUUCGACUCCAUACUGUCAUC---GGUCUCCUUGCCAUCGGAAUCAUCGCCG---C------UUUCCAGCGAAUCUUUGUCGCAGGCGAUGGUCUGAACCACAUCAGACUCAUCCGAUGGCACG -----------------------------...(((((-.((.(((((.(((..(...---.)..))).))))))))))))....---........((((((((((..((((((((---.------....).((((.......)))).)))))))((((((......))))))...)))))))))).. ( -51.20, z-score = -3.93, R) >droSec1.super_20 876890 142 - 1148123 -----------------------------AUCAUAGU-CAUCGGAGUAGAAAGAUAG---GUAGUUCGACUCCAUACUGUCAUC---GGUCUCCUCGUCAUCGGAAUCAUCGCCG---C------UUUCCAGCGAAUCUUUGUCGCAGGCGAUGGUCUGAACCACAUCAGACUCAUCCGAUGGCACG -----------------------------........-.....(((.(((..(((.(---..(((..(....)..)))..))))---..))).)))(((((((((..((((((((---.------....).((((.......)))).)))))))((((((......))))))...)))))))))... ( -50.50, z-score = -3.32, R) >droYak2.chrX 8516781 109 - 21770863 -----------------------------AUCAUAGU-CAUCGUCGUAGAAGGAUAG---AUUAUCCCACUC---------------------------GUCGAAA------------C------UGCCCAGUGAAUCUGCGUCGCCGACGAUGGUCUCAGCCACAUCAGAACCAUCCGAUGACUCC -----------------------------.....(((-(((((.((((((.(((((.---..)))))((((.---------------------------(.((...------------.------)).).))))..)))))).)......((((((((.(......).)).)))))).))))))).. ( -31.00, z-score = -1.61, R) >droEre2.scaffold_4690 6078874 145 + 18748788 -----------------------------AUCAUAGC-CAUCGGAGUAGAAAGAUAG---AUUGUUCCACUCCUUGUCGUCGCU---UUCAUUCUUGCUGUCGGAAUCGUCGUCCAAGC------UACCCAAUGAAUCUUCGUCGCCGACGAUGAUCUUAUAGACAUCAGAAGCACCCAAUGCCACC -----------------------------......((-....(((((.(((.((...---.)).))).)))))........(((---((......((((((.(((.((((((((...((------.((.............)).)).))))))))))).)))).))...))))).......)).... ( -30.52, z-score = 0.25, R) >droAna3.scaffold_13248 1974454 174 - 4840945 UCACAGCCAUCGCAGUGGCCAGCGUCCAUCUCGAAGUCCAGCUCGGUGGCCACGUUG-CCAUCUUCGAGAUUAUCGCUGCCCUU---GGACGCCGGCUGGCUAAAGAUACUGUCC---C------CGGCCAGGGAUUCGUCAUCACCCACAAGGGUCUGGUCCAUGUCGGAGCUAUCCGACGGCACA .....(((......((((((((((...(((((((((.((.....))((((......)-))).)))))))))...))))((((((---(...(((((..(((..........))).---)------))))..(((...........))).)))))))..)).))))((((((....)))))))))... ( -70.90, z-score = -2.18, R) >droWil1.scaffold_180588 73457 157 - 1294757 -----------------------------AUUAUCGC-CCUUAUCGUGAGAACAUAGAGUGAUAACUGAAUCAUUAUCACCAGCUAAGGUCUGUUCCAUAUCGGAACCUUCGCUGGGACAUAGAGUGAUAAAUGAAUCAUUAUCACUGACUAUGGUCUGUUCCUUAUCGGAACUAUCGGAAAAAAUA -----------------------------.(((((((-.((..((....))....)).)))))))((((.......((.(((((.((((((((........))).))))).)))))))(((((((((((((........))))))))..)))))....(((((.....)))))..))))........ ( -49.30, z-score = -3.91, R) >consensus _____________________________AUCAUAGU_CAUCGGAGUAGAAAGAUAG___GUAAUUCGACUCCUUACUGUCAUC___GGUCUCCUUGCCGUCGGAAUCAUCGCCG___C______UGCCCAGCGAAUCUUUGUCGCAGACGAUGGUCUGAUCCACAUCAGAACCAUCCGAUGGCACC ........................................((......))................................................(((((((..........................((((.......)))).......(((((((......))))).)).)))))))..... ( -7.01 = -7.49 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:05 2011