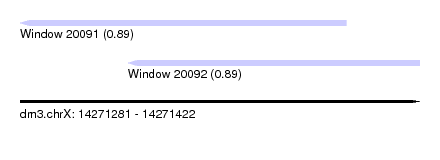
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,271,281 – 14,271,422 |
| Length | 141 |
| Max. P | 0.892914 |

| Location | 14,271,281 – 14,271,396 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.50248 |
| G+C content | 0.49443 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -22.21 |
| Energy contribution | -25.35 |
| Covariance contribution | 3.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
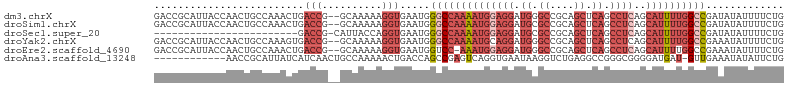
>dm3.chrX 14271281 115 - 22422827 CCGGCAAAAAGGUGAAUGGGCCAAAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGGAG-AAGACUCCAG--- ..(((...((((((.((.((((((((((((((.(((((....))))).))))..)))))))))).)).)))))).((((....)))).......)))....((((-....))))..--- ( -44.90, z-score = -3.16, R) >droSim1.chrX 10964690 117 - 17042790 CCGGCAAAAAGGUGAAUGGGCCAAAAUGGAGGAUGCGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGGAG--AGACACCACCAG ..........((((.((.((((((((((((((.((.((....)).)).))))..)))))))))).))...((((((.....(((.........))).....))))--)).))))..... ( -37.90, z-score = -1.50, R) >droSec1.super_20 874744 107 - 1148123 ----------GGUGAAUGGGCCAAAAUGGAGGAUGCGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGGAG--AGACACCACCAG ----------((((.((.((((((((((((((.((.((....)).)).))))..)))))))))).))...((((((.....(((.........))).....))))--)).))))..... ( -37.70, z-score = -2.35, R) >droYak2.chrX 8514530 116 - 21770863 CCGGCAAAAAGGUGAAUGGGCCAAAAUGCAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAAAAAGGAGAGACAUGAA--- ..(((.......((((((((((((((((((((.(((((....))))).)))..)))))))))))...........((((....)))))))))).)))...................--- ( -38.60, z-score = -1.66, R) >droEre2.scaffold_4690 6076606 107 + 18748788 CCGGCAAAAAGGUGAAUGGUCC-AAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGAAG--AGA--------- ..(((....(((((..(((((.-(((((((((.(((((....))))).))))..))))).)))))..........((((....)))))))))..)))........--...--------- ( -30.80, z-score = -0.04, R) >droAna3.scaffold_13248 1971038 101 - 4840945 ------------CCAGCCGAGUCAGGUGAAUAA-GGUCUGAGGCCGGGC---GGGGAUGAUGUUGAAAUAUAUUCUGCAAUGUUGCACAUUUAUACAAGAGAGUGAGAGAGAGACAG-- ------------((.(((...(((((.......-..)))))))).))((---((((...((((....)))).))))))..((((.(.(((((........))))).).....)))).-- ( -18.70, z-score = 1.06, R) >consensus CCGGCAAAAAGGUGAAUGGGCCAAAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGGAG__AGACACCAA___ ..........((((((((((((((((((((((.(((((....))))).))))..))))))))))...........((((....)))))))))..)))...................... (-22.21 = -25.35 + 3.14)

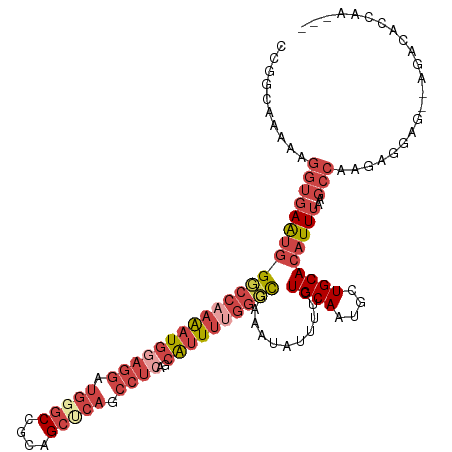
| Location | 14,271,319 – 14,271,422 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 70.59 |
| Shannon entropy | 0.55653 |
| G+C content | 0.50946 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -13.65 |
| Energy contribution | -16.07 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14271319 103 - 22422827 GACCGCAUUACCAACUGCCAAACUGACCG--GCAAAAAGGUGAAUGGGCCAAAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUG .......(((((...((((.........)--)))....)))))((.((((((((((((((.(((((....))))).))))..)))))))))).)).......... ( -37.90, z-score = -2.80, R) >droSim1.chrX 10964730 103 - 17042790 GACCGCAUUACCAACUGCCAAACUGACCG--GCAAAAAGGUGAAUGGGCCAAAAUGGAGGAUGCGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUG .......(((((...((((.........)--)))....)))))((.((((((((((((((.((.((....)).)).))))..)))))))))).)).......... ( -34.30, z-score = -2.03, R) >droSec1.super_20 874784 80 - 1148123 ------------------------GACCG-CAUUACCAGGUGAAUGGGCCAAAAUGGAGGAUGCGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUG ------------------------.(((.-........)))..((.((((((((((((((.((.((....)).)).))))..)))))))))).)).......... ( -27.30, z-score = -1.65, R) >droYak2.chrX 8514569 103 - 21770863 GACCGCAUUACCAACUGCCAAAGUGACCG--GCAAAAAGGUGAAUGGGCCAAAAUGCAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUG .....(((((((...((((.........)--)))....)))).)))((((((((((((((.(((((....))))).)))..)))))))))))............. ( -37.80, z-score = -2.51, R) >droEre2.scaffold_4690 6076637 102 + 18748788 GACCGCAUUACCAACUGCCAAACUGACCG--GCAAAAAGGUGAAUGGUCC-AAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUG .......(((((...((((.........)--)))....))))).(((((.-(((((((((.(((((....))))).))))..))))).)))))............ ( -30.50, z-score = -0.97, R) >droAna3.scaffold_13248 1971078 92 - 4840945 ------------AACCGCAUUAUCAUCAACUGCCAAAAACUGACCAGCCGAGUCAGGUGAAUAAGGUCUGAGGCCGGGCGGGGAUGAU-GUUGAAAUAUAUUCUG ------------..(.((((((((.....(((((.....((((((....).)))))........(((.....))).))))).))))))-)).)............ ( -27.30, z-score = -1.50, R) >consensus GACCGCAUUACCAACUGCCAAACUGACCG__GCAAAAAGGUGAAUGGGCCAAAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUG ..............................................((((((((((((((.((.((....)).)).))))..))))))))))............. (-13.65 = -16.07 + 2.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:04 2011