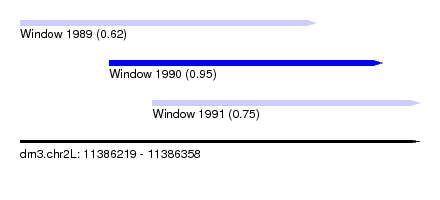
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,386,219 – 11,386,358 |
| Length | 139 |
| Max. P | 0.949505 |

| Location | 11,386,219 – 11,386,322 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.79 |
| Shannon entropy | 0.57723 |
| G+C content | 0.54832 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.24 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
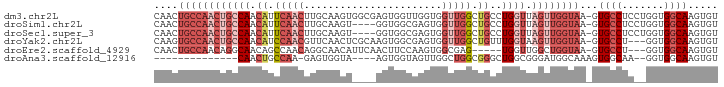
>dm3.chr2L 11386219 103 + 23011544 ------UCACCGAAUCUUAUCUGUGCAUCUCGCGGCAACGCCAGGUUGUCGCCAACUGCCAACUGCCAACAUUCAACU-------UGCAAGUGGCGAGUGGUUGGUGGUUGGCUGC ------.(((.((......)).)))......((((((((.....)))))))).....((((((..(((((.....(((-------(((.....)))))).)))))..))))))... ( -43.50, z-score = -3.18, R) >droSim1.chr2L 11194336 99 + 22036055 ------UCACCGAAUCUUAUCUGUGCAUCUCGCGGCAACGCCAGGUUGUCGCCAACUGCCAACUGCCAACAUUCAACU-------UGCAAGU----GGUGGCGAGUGGUUGGCUGC ------...............((.(((....((((((((.....))))))))....)))))...((((((.....(((-------(((....----....)))))).))))))... ( -32.30, z-score = -0.47, R) >droSec1.super_3 6780591 99 + 7220098 ------UCACCGAAUCUUAUCUGUGCAUCUCGCGGCAACGCCAGGUUGUCGCCAACUGCCAACUGCCAACAUUCAACU-------UGCAAGU----GGUGGCGAGUGGUUGGCUGC ------...............((.(((....((((((((.....))))))))....)))))...((((((.....(((-------(((....----....)))))).))))))... ( -32.30, z-score = -0.47, R) >droYak2.chr2L 7783711 102 + 22324452 ------UCACCGAAUCUUAUCUGUGCAUCU-GCGGCAACGCCAGGUUGUCGCCAAGUGCCAACUGCCAACAUCCAACG-------UUCAACUCGCAAGUGGCGAGUGGUUGGCUGU ------...............((.((((..-((((((((.....))))))))...))))))((.((((((........-------....((((((.....)))))).)))))).)) ( -34.42, z-score = -1.20, R) >droEre2.scaffold_4929 12583443 106 - 26641161 ------UCACCGAAUCUUAUCUGUGCAUCU-GCGGCAACGCCAGGUUGUCGCCAACUGCCAACAGGCAACAGCCAACAGGCAACAUUCAACUUCCAAGUGGCGAGUGGUUGGC--- ------..(((((((.((..((((......-(((....)))..((((((.(((...........))).)))))).))))..)).)))).((((((....)).)))))))....--- ( -33.30, z-score = -0.69, R) >droAna3.scaffold_12916 12094800 90 - 16180835 UCACCGAAUCCAAAUCUUAUCUGUGCAUCUCGAGGCAACGCCAGGUUGUCGCCAACUGCCAAGAG-UGGUAAGUGGUA-------GUUGGCUGGCGGG------------------ ...(((...(((.....................((((((.....))))))(((((((((((....-.......)))))-------)))))))))))).------------------ ( -30.70, z-score = -1.34, R) >droWil1.scaffold_181038 346670 83 + 637489 -------CACCGAAUCUUAUCUGUGCAUCUCGUGGCAACGCCAAGUUGUCGCCAAGUGCCAAGUG-UGGCAAGUGGUG-------UGGAGUUGCAUGU------------------ -------...............(((((.(((..((((((.....))))))((((..(((((....-)))))..)))).-------..))).)))))..------------------ ( -31.20, z-score = -2.47, R) >droGri2.scaffold_15126 7205521 78 - 8399593 ----------AGAAUCUUAUCUGUGCAUCUCGUGGCAACGCCAAGUUGUCGCCAAAUGCCUA--G-UGGCAAGUCGCA-------AGUGGCAGCCGGG------------------ ----------..........(((.((.......((((((.....))))))((((..((((..--.-.))))...(...-------.))))).))))).------------------ ( -25.40, z-score = -1.02, R) >consensus ______UCACCGAAUCUUAUCUGUGCAUCUCGCGGCAACGCCAGGUUGUCGCCAACUGCCAACUGCCAACAUGCAACA_______UGCAACUGCCGGGUGGCGAGUGGUUGGC___ .....................((.(((..(.((((((((.....)))))))).)..)))))....................................................... (-14.14 = -14.24 + 0.09)

| Location | 11,386,250 – 11,386,345 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 72.05 |
| Shannon entropy | 0.53753 |
| G+C content | 0.56647 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -16.25 |
| Energy contribution | -19.23 |
| Covariance contribution | 2.98 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11386250 95 + 23011544 ACGCCAGGUUGUCGCCAACUGCCAACUGCCAACAUUCAACUUGCAAGUGGCGAGUGGUUGGUGG-UUGGCUGCCUGGUUAGUUGGUAAGUGCCUCC ..(((((((....((((((..((((((...........((((((.....))))))))))))..)-))))).))))))).....(((....)))... ( -41.40, z-score = -2.60, R) >droSim1.chr2L 11194367 91 + 22036055 ACGCCAGGUUGUCGCCAACUGCCAACUGCCAACAUUCAACUUGCAAGU----GGUGGCGAGUGG-UUGGCUGCCUGGUUAGUUGGUAAGUGCCUCC .....((((....(((((((((((...((((((.....((((((....----....)))))).)-)))))....))).))))))))....)))).. ( -35.90, z-score = -1.53, R) >droSec1.super_3 6780622 91 + 7220098 ACGCCAGGUUGUCGCCAACUGCCAACUGCCAACAUUCAACUUGCAAGU----GGUGGCGAGUGG-UUGGCUGCCUGGUUAGUUGGUAAGUGCCUCC .....((((....(((((((((((...((((((.....((((((....----....)))))).)-)))))....))).))))))))....)))).. ( -35.90, z-score = -1.53, R) >droYak2.chr2L 7783741 92 + 22324452 ACGCCAGGUUGUCGCCAAGUGCCAACUGCCAACAUCCAACGUUCAACUCGCAAGUGGCGAGUGG-UUGGCUGUUUGGUAAGUUGGUAAGUGCC--- ..(((.(((....)))...(((((((((((((((.(((((.....((((((.....)))))).)-)))).)).))))).)))))))).).)).--- ( -36.70, z-score = -2.55, R) >droEre2.scaffold_4929 12583473 87 - 26641161 ACGCCAGGUUGUCGCCAACUGCCAACAGGCAACAGCCAACAGGCAACAUUCAACUUCCAAGUGG-CGAG-----UGGUUGGCUGGUAAGUGCC--- ..(((((.(..((((((..((((....))))...(((....))).................)))-))).-----.).))))).(((....)))--- ( -28.90, z-score = -0.29, R) >droAna3.scaffold_12916 12094837 75 - 16180835 ACGCCAGGUUGUCGCCAACUGCCAA------------GAGUGGUAAGUGGUAGUUGGCUGGCGGGCUGGCGGGAUGGCAAAGUGGCA--------- .((((((.((((((((((((((((.------------..........))))))))))).))))).)))))).....((......)).--------- ( -38.30, z-score = -4.00, R) >consensus ACGCCAGGUUGUCGCCAACUGCCAACUGCCAACAUUCAACUUGCAAGU_GCAAGUGGCGAGUGG_UUGGCUGCCUGGUUAGUUGGUAAGUGCC___ ......(((....(((((((((((...(((((...(((.(((((............)))))))).)))))....)))).)))))))....)))... (-16.25 = -19.23 + 2.98)


| Location | 11,386,265 – 11,386,358 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 70.75 |
| Shannon entropy | 0.55619 |
| G+C content | 0.54479 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -12.02 |
| Energy contribution | -15.05 |
| Covariance contribution | 3.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11386265 93 + 23011544 CAACUGCCAACUGCCAACAUUCAACUUGCAAGUGGCGAGUGGUUGGUGGUUGGCUGCCUGGUUAGUUGGUAA-GUGCCUCCUGGUGGCAAGUGU ..(((((((((..(((((.....((((((.....)))))).)))))..))))))((((...((((..(((..-..)))..)))).))))))).. ( -41.10, z-score = -2.88, R) >droSim1.chr2L 11194382 89 + 22036055 CAACUGCCAACUGCCAACAUUCAACUUGCAAGU----GGUGGCGAGUGGUUGGCUGCCUGGUUAGUUGGUAA-GUGCCUCCUGGUGGCAAGUGU (((((((((...((((((.....((((((....----....)))))).))))))....))).)))))).((.-.((((.......))))..)). ( -32.50, z-score = -0.70, R) >droSec1.super_3 6780637 89 + 7220098 CAACUGCCAACUGCCAACAUUCAACUUGCAAGU----GGUGGCGAGUGGUUGGCUGCCUGGUUAGUUGGUAA-GUGCCUCCUGGUGGCAAGUGU (((((((((...((((((.....((((((....----....)))))).))))))....))).)))))).((.-.((((.......))))..)). ( -32.50, z-score = -0.70, R) >droYak2.chr2L 7783756 90 + 22324452 CAAGUGCCAACUGCCAACAUCCAACGUUCAACUCGCAAGUGGCGAGUGGUUGGCUGUUUGGUAAGUUGGUAA-GUGCCU---GGUGGCAAGUGU ....(((((((((((((((.(((((.....((((((.....)))))).))))).)).))))).)))))))).-.((((.---...))))..... ( -38.90, z-score = -3.59, R) >droEre2.scaffold_4929 12583488 85 - 26641161 CAACUGCCAACAGGCAACAGCCAACAGGCAACAUUCAACUUCCAAGUGGCGAG-----UGGUUGGCUGGUAA-GUGCCU---GGUGGCAAGUGU ((..(((((.((((((.((((((((.(....).....((((((....)).)))-----).))))))))....-.)))))---).)))))..)). ( -39.30, z-score = -3.47, R) >droAna3.scaffold_12916 12094852 73 - 16180835 --------------CAACUGCCAA-GAGUGGUA----AGUGGUAGUUGGCUGGCGGGCUGGCGGGAUGGCAAAGUGGCAA--GGUGGCAAGUGU --------------(((((((((.-........----..)))))))))((((.(..(((.((......)).....)))..--).))))...... ( -21.30, z-score = -0.83, R) >consensus CAACUGCCAACUGCCAACAUCCAACUUGCAAGU____AGUGGCGAGUGGUUGGCUGCCUGGUUAGUUGGUAA_GUGCCUC__GGUGGCAAGUGU ....((((((((((((.((.(((((.......................))))).))..)))).))))))))...((((.......))))..... (-12.02 = -15.05 + 3.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:21 2011