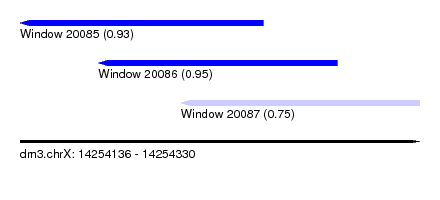
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,254,136 – 14,254,330 |
| Length | 194 |
| Max. P | 0.953791 |

| Location | 14,254,136 – 14,254,254 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Shannon entropy | 0.32645 |
| G+C content | 0.41954 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -20.52 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
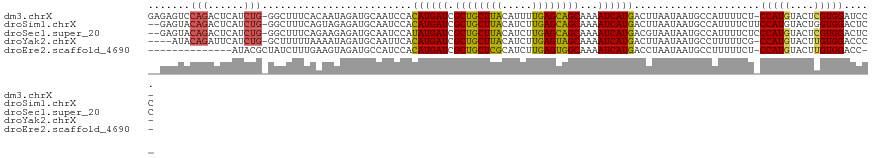
>dm3.chrX 14254136 118 - 22422827 GAGAGUCCAGACUCAUCUG-GGCUUUCACAAUAGAUGCAAUCCACAUGAUCGCUGCUUACAUUUUGAGCAGCAAAAUCAUGACUUAAUAAUGCCAUUUUCU-CCAUGUACUCGUGGAUCC- (((((((((((....))))-))))))).................((((((.((((((((.....))))))))...))))))...................(-(((((....))))))...- ( -40.20, z-score = -4.88, R) >droSim1.chrX 10948039 118 - 17042790 --GAGUACAGACUCAUCUG-GGCUUUCAGUAGAGAUGCAAUCCACAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUGACUUAAUAAUGCCAUUUUCUUCCAUGUACUGGUGGACUCC --((((.((((....))))-(((.....(((....)))......((((((.((((((((.....))))))))...))))))..........)))........((((......)))))))). ( -31.80, z-score = -1.51, R) >droSec1.super_20 856680 118 - 1148123 --GAGUACAGACUCAUCUG-GGCUUUCAGAAGAGAUGCAAUCCAUAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUGACGUAAUAAUGCCAUUUUCUCCCAUGUACUCGUGGACUCC --(((((((..(((.((((-(....))))).)))..........((((((.((((((((.....))))))))...))))))........................)))))))......... ( -32.40, z-score = -1.88, R) >droYak2.chrX 8497513 114 - 21770863 ----AUACAGAUUCAUCUG-GCUUUUUAAAAUAGAUGCAAUUCACAUGAUCGCUGCUUACAUCUUGAGUAGCAAAAUCAUGACUUAAUAAUGCCUUUUUCG-CCAUGUACUUGUGGACCC- ----.(((((...((..((-((...........((......)).((((((.((((((((.....))))))))...))))))...................)-)))))...))))).....- ( -23.50, z-score = -1.26, R) >droEre2.scaffold_4690 6060012 104 + 18748788 --------------AUACGCUAUCUUUGAAGUAGAUGCCAUCCACAUGAUCGCUGCUCGCAUCUUGAGUGGCAAAAUCAUGACCUAAUAAUGCCUUUUUCU-CCAUGUACUUGUGGACC-- --------------....((.((((.......))))))......((((((.((..((((.....))))..))...))))))...................(-((((......)))))..-- ( -23.20, z-score = -1.19, R) >consensus __GAGUACAGACUCAUCUG_GGCUUUCAAAAUAGAUGCAAUCCACAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUGACUUAAUAAUGCCAUUUUCU_CCAUGUACUCGUGGACCC_ ............................................((((((.((((((((.....))))))))...)))))).....................(((((....)))))..... (-20.52 = -19.68 + -0.84)
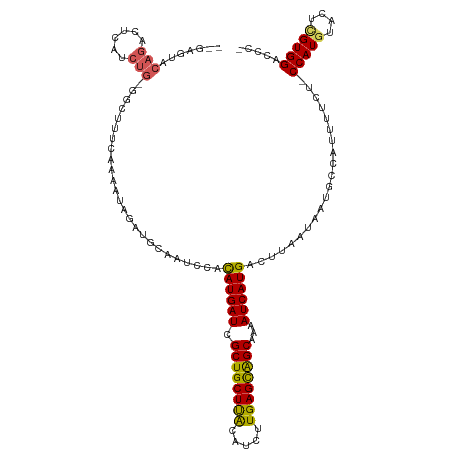
| Location | 14,254,174 – 14,254,290 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Shannon entropy | 0.38579 |
| G+C content | 0.39021 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -18.98 |
| Energy contribution | -17.50 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14254174 116 - 22422827 AAGAUCUUUUAUUAUC----AUAAGUAGCUCAGAGAGAUAGAGAGUCCAGACUCAUCUGGGCUUUCACAAUAGAUGCAAUCCACAUGAUCGCUGCUUACAUUUUGAGCAGCAAAAUCAUG ...(((((((......----............))))))).(((((((((((....))))))))))).................((((((.((((((((.....))))))))...)))))) ( -35.37, z-score = -3.47, R) >droSim1.chrX 10948079 112 - 17042790 UAGAUCUUUUAUUAUC----AUAAGUAACUCAGUGAG----AGAGUACAGACUCAUCUGGGCUUUCAGUAGAGAUGCAAUCCACAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUG ................----....(((.((((.((((----((....((((....))))..)))))).).))).)))......((((((.((((((((.....))))))))...)))))) ( -32.20, z-score = -2.35, R) >droSec1.super_20 856720 112 - 1148123 UAGACCUUUUAUUAUC----AUAAGCAGCUCAGUGAG----AGAGUACAGACUCAUCUGGGCUUUCAGAAGAGAUGCAAUCCAUAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUG ................----....(((.(((..(..(----((((..((((....))))..))))).)..))).)))......((((((.((((((((.....))))))))...)))))) ( -31.70, z-score = -1.80, R) >droYak2.chrX 8497551 112 - 21770863 AAGAUCUUUUAUUAUU--UAAUAAGUAGCUCAGCG------AGAAUACAGAUUCAUCUGGCUUUUUAAAAUAGAUGCAAUUCACAUGAUCGCUGCUUACAUCUUGAGUAGCAAAAUCAUG ..((.......(((((--(..((((.(((.(((..------.((((....))))..)))))).)))))))))).......)).((((((.((((((((.....))))))))...)))))) ( -23.04, z-score = -0.94, R) >droEre2.scaffold_4690 6060049 105 + 18748788 AAGAUCUCUUACUUUUAAGUACGAGUAUCUCAG--------CGGGCAUACGCU-------AUCUUUGAAGUAGAUGCCAUCCACAUGAUCGCUGCUCGCAUCUUGAGUGGCAAAAUCAUG .(((((((.((((....)))).))).))))...--------..(((((..(((-------........)))..))))).....((((((.((..((((.....))))..))...)))))) ( -31.40, z-score = -2.44, R) >consensus AAGAUCUUUUAUUAUC____AUAAGUAGCUCAGUGAG____AGAGUACAGACUCAUCUGGGCUUUCAAAAUAGAUGCAAUCCACAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUG ..........................................((((....)))).............................((((((.((((((((.....))))))))...)))))) (-18.98 = -17.50 + -1.48)

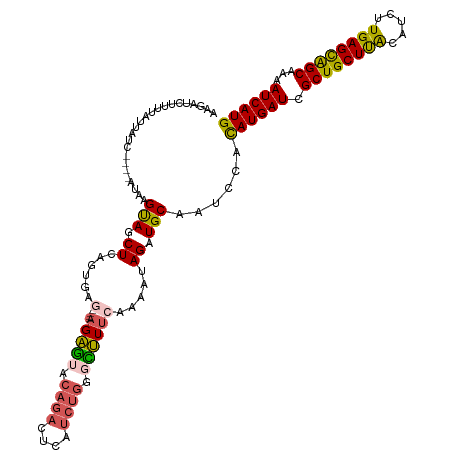
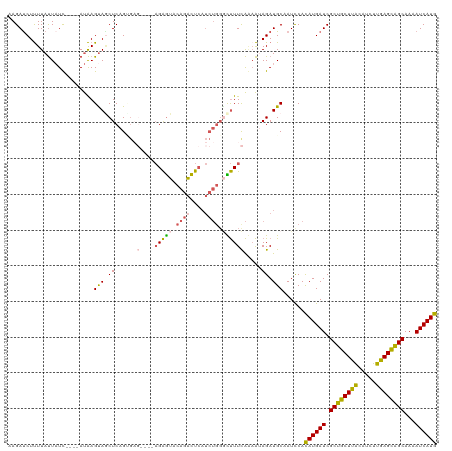
| Location | 14,254,214 – 14,254,330 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.69 |
| Shannon entropy | 0.42253 |
| G+C content | 0.32879 |
| Mean single sequence MFE | -24.79 |
| Consensus MFE | -14.84 |
| Energy contribution | -15.48 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747406 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 14254214 116 - 22422827 UUUAUUAAAUUGUGUGAAAACCAUUUACUUUGCAUUUCCCAAGAUCUUUUAUUAUC----AUAAGUAGCUCAGAGAGAUAGAGAGUCCAGACUCAUCUGGGCUUUCACAAUAGAUGCAAU .....(((((.(.((....))))))))..((((((((......(((((((......----............))))))).(((((((((((....))))))))))).....)))))))). ( -28.17, z-score = -2.46, R) >droSim1.chrX 10948119 112 - 17042790 UUUAUUAAAUUGUGUGUAAACCAUUUACUUUGCAUUUCCCUAGAUCUUUUAUUAUC----AUAAGUAACUCAGUG----AGAGAGUACAGACUCAUCUGGGCUUUCAGUAGAGAUGCAAU .....(((((.((......)).)))))..(((((((((................((----((..........)))----)(((((..((((....))))..)))))....))))))))). ( -23.90, z-score = -1.07, R) >droSec1.super_20 856760 112 - 1148123 AUUAUUAAAUUGUGUGUAAACCAUUUACUUUGCAUUUCCCUAGACCUUUUAUUAUC----AUAAGCAGCUCAGUG----AGAGAGUACAGACUCAUCUGGGCUUUCAGAAGAGAUGCAAU .....(((((.((......)).)))))..(((((((((....((.(((((((....----))))).)).))....----.(((((..((((....))))..)))))....))))))))). ( -24.60, z-score = -1.02, R) >droYak2.chrX 8497591 112 - 21770863 UUUAUUAAAUGUUGUGAAAAGCAUGUAUUUUGCAUUUACCAAGAUCUUUUAUUAUU--UAAUAAGUAGCUCAG------CGAGAAUACAGAUUCAUCUGGCUUUUUAAAAUAGAUGCAAU ........(((((......))))).....(((((((((....((.((((((((...--.)))))).)).))..------.(((((..((((....))))..)))))....))))))))). ( -19.20, z-score = -0.10, R) >droEre2.scaffold_4690 6060089 105 + 18748788 UUUAUUAAAUUGUAGGGAAACCAUUUAUUUAGCAUUUGCCAAGAUCUCUUACUUUUAAGUACGAGUAUCUC--------AGCGGGCAUACGCU-------AUCUUUGAAGUAGAUGCCAU .....(((((....((....))....)))))((((((((..(((((((.((((....)))).))).)))).--------((((......))))-------.........))))))))... ( -28.10, z-score = -3.08, R) >consensus UUUAUUAAAUUGUGUGAAAACCAUUUACUUUGCAUUUCCCAAGAUCUUUUAUUAUC____AUAAGUAGCUCAG_G____AGAGAGUACAGACUCAUCUGGGCUUUCAAAAUAGAUGCAAU .....(((((.((......)).)))))..(((((((((....((.(((((((........))))).)).)).........((((((.(((......))).))))))....))))))))). (-14.84 = -15.48 + 0.64)

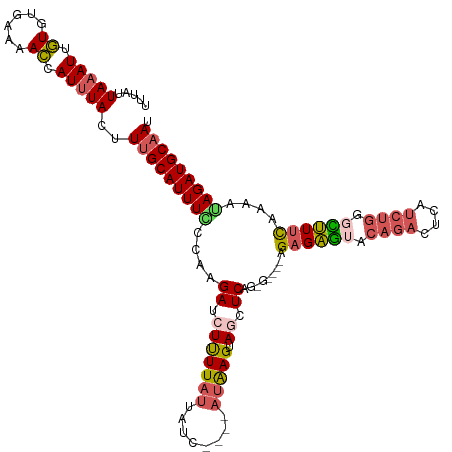
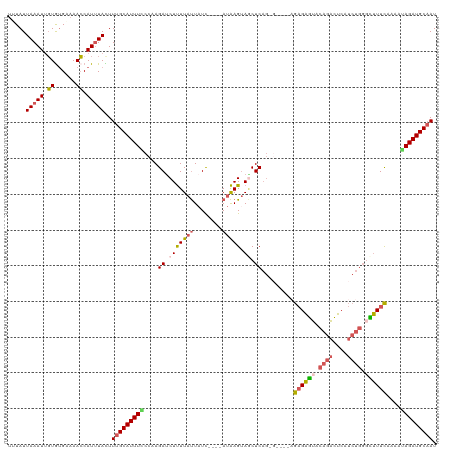
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:43:00 2011