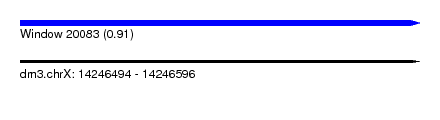
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,246,494 – 14,246,596 |
| Length | 102 |
| Max. P | 0.908613 |

| Location | 14,246,494 – 14,246,596 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.09 |
| Shannon entropy | 0.46127 |
| G+C content | 0.52927 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -16.45 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14246494 102 + 22422827 CUGCGGUCCAUCAGGAUCAACGUUUUGGU--AUUAAUUAAUGAUGCCGAACGUCCUGCCGUCCUGAUGUCCGCCGUCCGCCGU-------CCGCUG-UCCUUUG-UCCUUUUG ..((((..(((((((((((((((((.(((--((((.....)))))))))))))..))..))))))))).)))).....((...-------..))..-.......-........ ( -31.80, z-score = -3.85, R) >droAna3.scaffold_13248 1940297 93 + 4840945 CUGUUCUCCAUCAGGAUCAACGUUUUGGGGGAUUAAUUAAUGAUGAUGGACAG---GAGGACUGGACGGAAAGGACACCCGGA--------CAGUGUCCUUUCA--------- ((((((..(((((.((((..(......)..))))......)))))..))))))---............((((((((((.....--------..)))))))))).--------- ( -31.90, z-score = -1.87, R) >droEre2.scaffold_4690 6052351 111 - 18748788 CUGCGGUCCAUCAGGAUCAACGUUUUGGG--AUUAAUUAAUGAUGCCAAACGUCCUGCAGUCCUGUUGUCCACCAUCCGCCAGCCGCCAUCCUCUGGCCCUCUGCUCCAUUUG ..(((((.((.((((((((((((((.((.--((((.....)))).))))))))..))..)))))).))...)))....(((((..........))))).....))........ ( -28.50, z-score = -1.05, R) >droYak2.chrX 8489995 109 + 21770863 CUGCGGUCCAUCAGGAUCAACGUUUUGGG--AUUAAUUAAUGAUGCCGAACGUCCUACAGUCCUGCUGUCCGCCAUUCGCCAUCCGCCAUCCGCUA-UCCGCUG-UCCUUUUG ..((((..((.((((((..((((((.((.--((((.....)))).))))))))......)))))).)).)))).....((.....((.....))..-...))..-........ ( -24.90, z-score = -0.33, R) >droSec1.super_20 849174 109 + 1148123 CUGCGGUCCAUCAGGAUCAACGUUUUGGG--AUUAAUUAAUGAUGCCGAACGUCCUGCAGUCCUGAUGUCCGCCGUCCGCCGUCCGCUUUCCGCCG-UCCUUUG-UCCUUUUG ..((((..(((((((((((((((((.((.--((((.....)))).))))))))..))..))))))))).)))).(..((.((.........)).))-..)....-........ ( -30.60, z-score = -1.79, R) >droSim1.chrX_random 3812726 102 + 5698898 UUGCGGUCCAUCAGGAUCAACGUUUUGGG--AUUAAUUAAUGAUGCCGAACGUCCUGCAGUCCUGAUGUCCGCCGUCCGCUGU-------CCGCCG-UCCUUUG-UCCUUUUG ..((((..(((((((((((((((((.((.--((((.....)))).))))))))..))..))))))))).)))).(.(.((...-------..)).)-.).....-........ ( -29.70, z-score = -1.98, R) >consensus CUGCGGUCCAUCAGGAUCAACGUUUUGGG__AUUAAUUAAUGAUGCCGAACGUCCUGCAGUCCUGAUGUCCGCCAUCCGCCGU_______CCGCUG_UCCUUUG_UCCUUUUG ..(((((.(((((((((..((((((.((...((((.....)))).))))))))......)))))))))...)))))..................................... (-16.45 = -16.48 + 0.03)

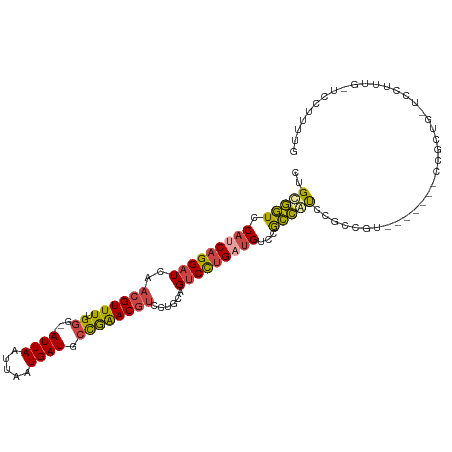

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:56 2011