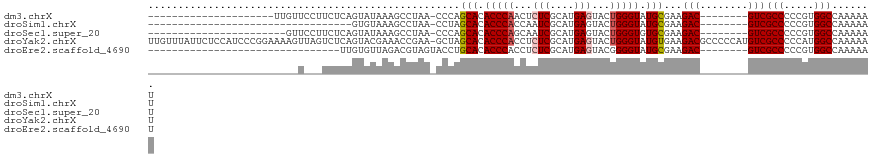
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,236,413 – 14,236,504 |
| Length | 91 |
| Max. P | 0.979321 |

| Location | 14,236,413 – 14,236,504 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 70.15 |
| Shannon entropy | 0.45307 |
| G+C content | 0.53163 |
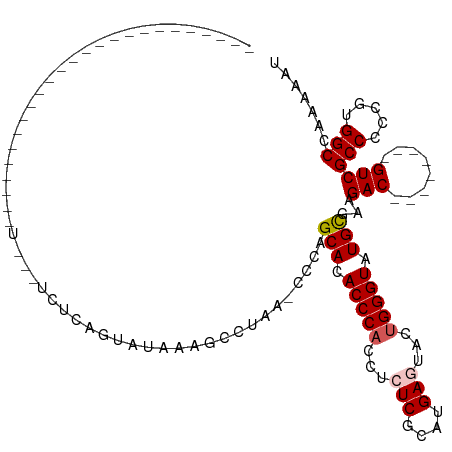
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -25.02 |
| Energy contribution | -25.46 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
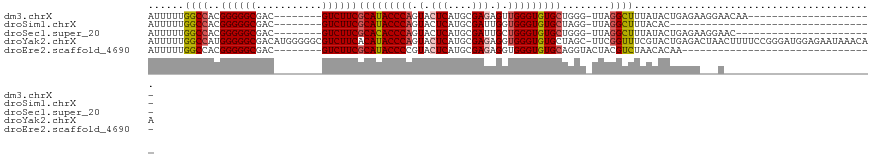
>dm3.chrX 14236413 91 + 22422827 AUUUUUGGCCACGGGGGCGAC--------GUCUUCGCAUACCCAGUACUCAUGCGAGAGUUGGGUGUGCUGGG-UUAGGCUUUAUACUGAGAAGGAACAA--------------------- .......(((.....)))...--------.(((((((((((((((..(((....)))..))))))))))....-((((........))))))))).....--------------------- ( -31.20, z-score = -1.75, R) >droSim1.chrX 10930863 78 + 17042790 AUUUUUGGCCACGGGGGCGAC--------GUCUUCGCAUACCCAGUACUCAUGCGAUUGGUGGGUGUGCUAGG-UUAGGCUUUACAC---------------------------------- ...(((((((..((((((...--------))))))(((((((((.((.((....)).)).)))))))))..))-)))))........---------------------------------- ( -28.60, z-score = -2.38, R) >droSec1.super_20 839180 89 + 1148123 AUUUUUGGCCACGGGGGCGAC--------GUCUUCGCACACCCAGUACUCAUGCGAUUGCUGGGUGUGCUGGG-UUAGGCUUUAUACUGAGAAGGAAC----------------------- .......(((.....)))...--------.(((((((((((((((((.((....)).))))))))))))....-((((........)))))))))...----------------------- ( -35.00, z-score = -3.09, R) >droYak2.chrX 8479284 120 + 21770863 AUUUUUGGCCAUGGGGGCGACAUGGGGGCGUCUUCACAUACCCAGUACUCAUGCGAGAGGUGGGUGUGCUAGC-UUCGGUUUCGUACUGAGACUAACUUUUCCGGGAUGGAGAAUAAACAA ((((((.(((.(((..(.(.....(((((.......((((((((.(.(((....))).).))))))))...))-)))((((((.....))))))..).)..))))).).))))))...... ( -35.70, z-score = -0.14, R) >droEre2.scaffold_4690 6043199 81 - 18748788 AUUUUUGGCCACGGGGGCGAC--------GUCUUCGCAUACCCCGUACUCAUGCGAGAGGUGGGUGUGCAGGUACUACGUCUAACACAA-------------------------------- .......(((.....)))(((--------(((((.(((((((((.(.(((....)))).).)))))))))))....)))))........-------------------------------- ( -28.00, z-score = -1.06, R) >consensus AUUUUUGGCCACGGGGGCGAC________GUCUUCGCAUACCCAGUACUCAUGCGAGAGGUGGGUGUGCUGGG_UUAGGCUUUAUACUGAGA___A_________________________ ......((((..((((((...........))))))(((((((((.(.(((....))).).)))))))))........))))........................................ (-25.02 = -25.46 + 0.44)
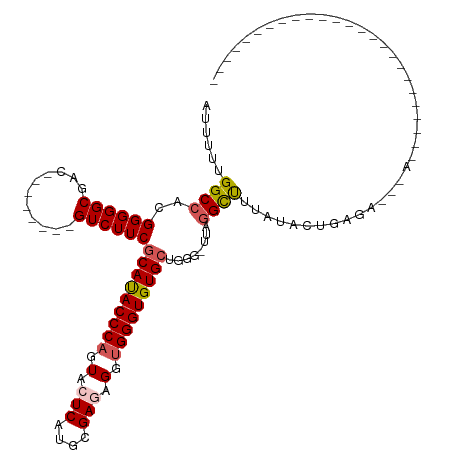
| Location | 14,236,413 – 14,236,504 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 70.15 |
| Shannon entropy | 0.45307 |
| G+C content | 0.53163 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -15.92 |
| Energy contribution | -16.56 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14236413 91 - 22422827 ---------------------UUGUUCCUUCUCAGUAUAAAGCCUAA-CCCAGCACACCCAACUCUCGCAUGAGUACUGGGUAUGCGAAGAC--------GUCGCCCCCGUGGCCAAAAAU ---------------------......((((...((.....))....-....(((.(((((((((......))))..))))).)))))))..--------(((((....)))))....... ( -22.50, z-score = -1.81, R) >droSim1.chrX 10930863 78 - 17042790 ----------------------------------GUGUAAAGCCUAA-CCUAGCACACCCACCAAUCGCAUGAGUACUGGGUAUGCGAAGAC--------GUCGCCCCCGUGGCCAAAAAU ----------------------------------.......(((...-....(((.(((((....((....))....))))).)))....((--------(.......))))))....... ( -16.50, z-score = -0.05, R) >droSec1.super_20 839180 89 - 1148123 -----------------------GUUCCUUCUCAGUAUAAAGCCUAA-CCCAGCACACCCAGCAAUCGCAUGAGUACUGGGUGUGCGAAGAC--------GUCGCCCCCGUGGCCAAAAAU -----------------------....((((...((.....))....-....((((((((((...((....))...))))))))))))))..--------(((((....)))))....... ( -27.40, z-score = -3.10, R) >droYak2.chrX 8479284 120 - 21770863 UUGUUUAUUCUCCAUCCCGGAAAAGUUAGUCUCAGUACGAAACCGAA-GCUAGCACACCCACCUCUCGCAUGAGUACUGGGUAUGUGAAGACGCCCCCAUGUCGCCCCCAUGGCCAAAAAU ..........((((((((((....(((((..((.((.....)).)).-.)))))........(((......)))..))))).))).)).((((......))))(((.....)))....... ( -27.50, z-score = -0.99, R) >droEre2.scaffold_4690 6043199 81 + 18748788 --------------------------------UUGUGUUAGACGUAGUACCUGCACACCCACCUCUCGCAUGAGUACGGGGUAUGCGAAGAC--------GUCGCCCCCGUGGCCAAAAAU --------------------------------........(((((......((((.((((....(((....)))....)))).))))...))--------)))(((.....)))....... ( -23.40, z-score = -0.65, R) >consensus _________________________U___UCUCAGUAUAAAGCCUAA_CCCAGCACACCCACCUCUCGCAUGAGUACUGGGUAUGCGAAGAC________GUCGCCCCCGUGGCCAAAAAU ....................................................(((.(((((...(((....)))...))))).)))...((((......))))(((.....)))....... (-15.92 = -16.56 + 0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:51 2011