
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,226,563 – 14,226,659 |
| Length | 96 |
| Max. P | 0.986433 |

| Location | 14,226,563 – 14,226,659 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 69.18 |
| Shannon entropy | 0.60594 |
| G+C content | 0.55315 |
| Mean single sequence MFE | -32.81 |
| Consensus MFE | -17.05 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.59 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
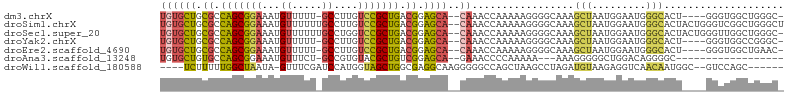
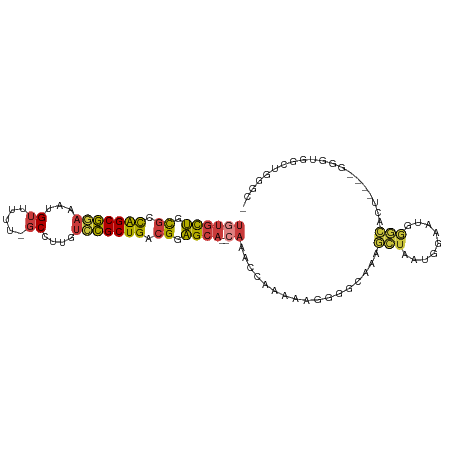
>dm3.chrX 14226563 96 + 22422827 UGUGCUGCGCCAGCGGAAAUGUUUUU-GCCUUGUCCGCUGACGGAGCA--CAAACCAAAAAGGGGCAAAGCUAAUGGAAUGGGCACU----GGGUGGCUGGGC- ((((((.((.(((((((...((....-))....))))))).)).))))--))..((......))((..(((((.(((........))----)..)))))..))- ( -34.80, z-score = -0.92, R) >droSim1.chrX 10921024 102 + 17042790 UGUGCUGCGCCAGCGGAAAUGUUUUUUGCCUUGUCCGCUGACGGAGCA--CAAACCAAAAAGGGGCAAAGCUAAUGGAAUGGGCACUACUGGGUCGGCUGGGCU ((((((.((.(((((((...((.....))....))))))).)).))))--))..((.....))(((..((((...((..(((......)))..))))))..))) ( -36.60, z-score = -0.83, R) >droSec1.super_20 829661 101 + 1148123 UGUGCUGCGCCAGCGGAAAUGUUUUUUGCCUGGUCCGCUGACGGAGCA--CAAACCAAAAAGGGGCAAAGCUAAUGGAAUGGGCACUACUGGGUUGGCUGGGC- ((((((.((.(((((((...((.....))....))))))).)).))))--))..((......))((..(((((((....(((......))).)))))))..))- ( -36.90, z-score = -1.15, R) >droYak2.chrX 8469721 96 + 21770863 UGUGCUGCGCCAGCGGAAAUGUUUUU-GCCUUGUCCGCUGACGGAGCA--CAAACCAAAAAGGGGCAAAGCUAAUGGAAUGGGCACU----GGGUGGCCGGGC- ((((((.((.(((((((...((....-))....))))))).)).))))--))..(((...(..(((...)))..)....)))((.((----((....))))))- ( -35.30, z-score = -0.70, R) >droEre2.scaffold_4690 6033311 96 - 18748788 UGUGCUGCGCCAGCGGAAAUGUUUUU-GCCUUGUCCGCUGACGGAGCA--CAAACCAAAAAGGGGCAAAGCUAAUGGAAUGGGCACU----GGGUGGCUGAAC- ((((((.((.(((((((...((....-))....))))))).)).))))--))..(((...((..((....(.....).....)).))----...)))......- ( -32.00, z-score = -0.74, R) >droAna3.scaffold_13248 1918728 80 + 4840945 UGUGCUGUGCCAGCGGAAAUGUUUCU-GCCGUGUACGCUGUCGGAGCA--GAAACCCCAAAAA---AAAGGGGGCUGGACAGGGGC------------------ ....((((.(((((.......(((((-((..((........))..)))--))))((((.....---...)))))))))))))....------------------ ( -30.20, z-score = -1.59, R) >droWil1.scaffold_180588 28641 91 + 1294757 ----UCUUUUUGGCUAAUA-GUUUCGAUCCAUGGUAGCUGGCGAGGCAAGGGGGCCAGCUAAGCCUAGAUGUAAGAGGUCAACAAUGGC--GUCCAGC------ ----........(((....-((((((..(((.......)))))))))..(((.((((.....((((.........))))......))))--.))))))------ ( -23.90, z-score = 0.87, R) >consensus UGUGCUGCGCCAGCGGAAAUGUUUUU_GCCUUGUCCGCUGACGGAGCA__CAAACCAAAAAGGGGCAAAGCUAAUGGAAUGGGCACU____GGGUGGCUGGGC_ ...(((.((.(((((((...((.....))....))))))).)).)))......................(((.........))).................... (-17.05 = -16.43 + -0.62)
| Location | 14,226,563 – 14,226,659 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 69.18 |
| Shannon entropy | 0.60594 |
| G+C content | 0.55315 |
| Mean single sequence MFE | -28.89 |
| Consensus MFE | -16.49 |
| Energy contribution | -17.51 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14226563 96 - 22422827 -GCCCAGCCACCC----AGUGCCCAUUCCAUUAGCUUUGCCCCUUUUUGGUUUG--UGCUCCGUCAGCGGACAAGGC-AAAAACAUUUCCGCUGGCGCAGCACA -((..(((.....----((((.......)))).)))..)).((.....))..((--((((.((((((((((...(..-.....)...)))))))))).)))))) ( -28.80, z-score = -1.27, R) >droSim1.chrX 10921024 102 - 17042790 AGCCCAGCCGACCCAGUAGUGCCCAUUCCAUUAGCUUUGCCCCUUUUUGGUUUG--UGCUCCGUCAGCGGACAAGGCAAAAAACAUUUCCGCUGGCGCAGCACA .....((((((....((((.((...........)).))))......))))))((--((((.((((((((((...(........)...)))))))))).)))))) ( -32.30, z-score = -1.82, R) >droSec1.super_20 829661 101 - 1148123 -GCCCAGCCAACCCAGUAGUGCCCAUUCCAUUAGCUUUGCCCCUUUUUGGUUUG--UGCUCCGUCAGCGGACCAGGCAAAAAACAUUUCCGCUGGCGCAGCACA -....((((((....((((.((...........)).))))......))))))((--((((.((((((((((...(........)...)))))))))).)))))) ( -32.60, z-score = -2.35, R) >droYak2.chrX 8469721 96 - 21770863 -GCCCGGCCACCC----AGUGCCCAUUCCAUUAGCUUUGCCCCUUUUUGGUUUG--UGCUCCGUCAGCGGACAAGGC-AAAAACAUUUCCGCUGGCGCAGCACA -....((.(((..----.))).))...(((..((........))...)))..((--((((.((((((((((...(..-.....)...)))))))))).)))))) ( -30.90, z-score = -1.28, R) >droEre2.scaffold_4690 6033311 96 + 18748788 -GUUCAGCCACCC----AGUGCCCAUUCCAUUAGCUUUGCCCCUUUUUGGUUUG--UGCUCCGUCAGCGGACAAGGC-AAAAACAUUUCCGCUGGCGCAGCACA -....(((((...----((.(..((............))..)))...)))))((--((((.((((((((((...(..-.....)...)))))))))).)))))) ( -28.50, z-score = -1.45, R) >droAna3.scaffold_13248 1918728 80 - 4840945 ------------------GCCCCUGUCCAGCCCCCUUU---UUUUUGGGGUUUC--UGCUCCGACAGCGUACACGGC-AGAAACAUUUCCGCUGGCACAGCACA ------------------....(((((((((.(((...---.....)))(((((--((((...((...))....)))-))))))......))))).)))).... ( -29.00, z-score = -3.14, R) >droWil1.scaffold_180588 28641 91 - 1294757 ------GCUGGAC--GCCAUUGUUGACCUCUUACAUCUAGGCUUAGCUGGCCCCCUUGCCUCGCCAGCUACCAUGGAUCGAAAC-UAUUAGCCAAAAAGA---- ------(((((((--(....))))..........(((((((..((((((((...........)))))))))).)))))......-...))))........---- ( -20.10, z-score = 0.06, R) >consensus _GCCCAGCCACCC____AGUGCCCAUUCCAUUAGCUUUGCCCCUUUUUGGUUUG__UGCUCCGUCAGCGGACAAGGC_AAAAACAUUUCCGCUGGCGCAGCACA ........................................................((((.((((((((((...(........)...)))))))))).)))).. (-16.49 = -17.51 + 1.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:48 2011