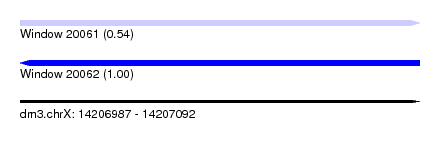
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,206,987 – 14,207,092 |
| Length | 105 |
| Max. P | 0.997935 |

| Location | 14,206,987 – 14,207,092 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.69 |
| Shannon entropy | 0.21408 |
| G+C content | 0.41854 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
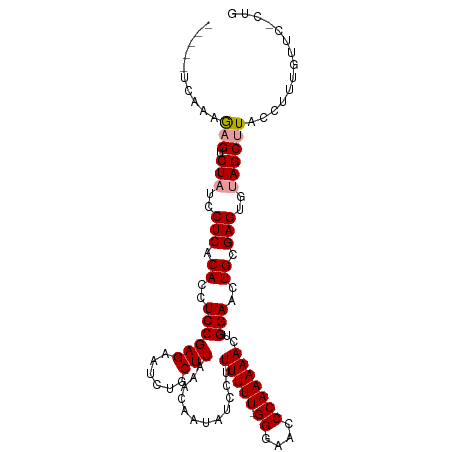
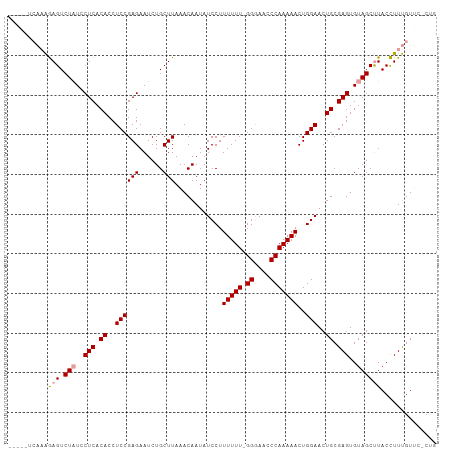
>dm3.chrX 14206987 105 + 22422827 --UUUUGUUGAAGUCUUUCCUCACACCUCCGAGAAUCUGCUUAAACAAUAUCCUUUUUU-GGAAAACCAAAAACUGGAACUGCGAGUGUAGCCUACUUUCGUAGGCUG --..(((((.((((.....(((........))).....)))).)))))..(((.(((((-((....)))))))..)))..........((((((((....)))))))) ( -27.00, z-score = -2.81, R) >droSec1.super_20 802816 102 + 1148123 -----ACAAAGAGUCUAUCCUCACACCUCCGAGAAUCUGCUUAAACAAUAUCCUUUUUUUGGGAACCCAAAAACUGGAACUGCGAGUGUAGCUUACCUUUGUUC-CUG -----((((((((.(((..(((.((.....(((......)))........(((..(((((((....)))))))..)))..)).)))..)))))...))))))..-... ( -23.70, z-score = -2.40, R) >droSim1.chrX_random 3805940 106 + 5698898 ACAGCUCAAAGAGUCUAUGCUCACACCUCCGAGAAUCUGCUUAAACAAUAUCCUUUUUU-GGGAACCCAAAAACUGGAACUGCGAGUGUAGCUUACCUUUGUUC-CUA ..((..(((((((.((((((((.((..((((((......)))............(((((-((....)))))))..)))..)).))))))))))...)))))...-)). ( -25.80, z-score = -2.12, R) >consensus _____UCAAAGAGUCUAUCCUCACACCUCCGAGAAUCUGCUUAAACAAUAUCCUUUUUU_GGGAACCCAAAAACUGGAACUGCGAGUGUAGCUUACCUUUGUUC_CUG ..........(((.(((..(((.((..((((((......)))............(((((.((....)))))))..)))..)).)))..)))))).............. (-14.52 = -14.97 + 0.45)

| Location | 14,206,987 – 14,207,092 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.69 |
| Shannon entropy | 0.21408 |
| G+C content | 0.41854 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -24.18 |
| Energy contribution | -25.40 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.997935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
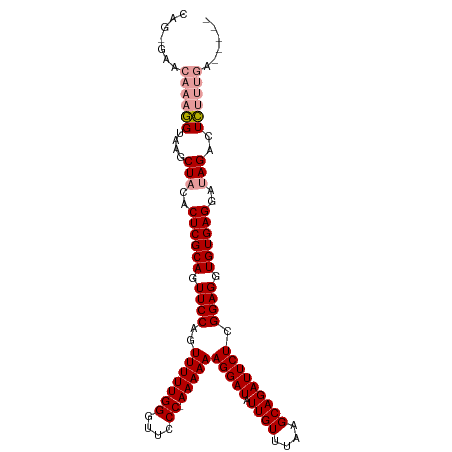
>dm3.chrX 14206987 105 - 22422827 CAGCCUACGAAAGUAGGCUACACUCGCAGUUCCAGUUUUUGGUUUUCC-AAAAAAGGAUAUUGUUUAAGCAGAUUCUCGGAGGUGUGAGGAAAGACUUCAACAAAA-- .(((((((....)))))))...((((((.((((..(((((((....))-)))))(((((.((((....))))))))).)))).)))))).................-- ( -39.90, z-score = -5.59, R) >droSec1.super_20 802816 102 - 1148123 CAG-GAACAAAGGUAAGCUACACUCGCAGUUCCAGUUUUUGGGUUCCCAAAAAAAGGAUAUUGUUUAAGCAGAUUCUCGGAGGUGUGAGGAUAGACUCUUUGU----- ...-..(((((((....(((..((((((.((((..(((((((....))))))).(((((.((((....))))))))).)))).))))))..)))..)))))))----- ( -35.40, z-score = -3.95, R) >droSim1.chrX_random 3805940 106 - 5698898 UAG-GAACAAAGGUAAGCUACACUCGCAGUUCCAGUUUUUGGGUUCCC-AAAAAAGGAUAUUGUUUAAGCAGAUUCUCGGAGGUGUGAGCAUAGACUCUUUGAGCUGU ..(-(..((((((....(((..((((((.((((..(((((((....))-)))))(((((.((((....))))))))).)))).))))))..)))..))))))..)).. ( -34.20, z-score = -2.77, R) >consensus CAG_GAACAAAGGUAAGCUACACUCGCAGUUCCAGUUUUUGGGUUCCC_AAAAAAGGAUAUUGUUUAAGCAGAUUCUCGGAGGUGUGAGGAUAGACUCUUUGA_____ .......((((((....(((..((((((.((((..(((((((....)).)))))(((((.((((....))))))))).)))).))))))..)))..))))))...... (-24.18 = -25.40 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:39 2011