
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,378,908 – 11,379,011 |
| Length | 103 |
| Max. P | 0.835033 |

| Location | 11,378,908 – 11,379,011 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Shannon entropy | 0.28082 |
| G+C content | 0.37400 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.62 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
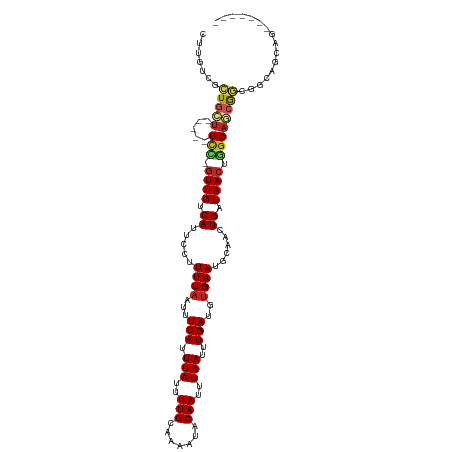
>dm3.chr2L 11378908 103 + 23011544 CUUGUCGCUGCU-----GCC-GUUGUCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUGGCAGCAGCGGCAGUAG------- .(((((((((((-----(((-(((((((.......(((......)))..(((((..((((((......))).)))...))))).))))))).))))))))))))))...------- ( -41.10, z-score = -6.48, R) >droEre2.scaffold_4929 12576129 103 - 26641161 CUUGUCGCUGCU-----GCC-GUUGUCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUGGCAGCAGCGGCAGCAG------- .(((((((((((-----(((-(((((((.......(((......)))..(((((..((((((......))).)))...))))).))))))).))))))))))))))...------- ( -41.00, z-score = -6.11, R) >droSec1.super_3 6773339 103 + 7220098 CUUGUCGCUGCU-----GCC-GUUGUCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUGGCAGCAGCGGCAGUAG------- .(((((((((((-----(((-(((((((.......(((......)))..(((((..((((((......))).)))...))))).))))))).))))))))))))))...------- ( -41.10, z-score = -6.48, R) >droSim1.chr2L 11185031 103 + 22036055 CUUGUCGCUGCU-----GCC-GUUGUCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUGGCAGCAGCGUCAGUAG------- .(((.(((((((-----(((-(((((((.......(((......)))..(((((..((((((......))).)))...))))).))))))).)))))))))).)))...------- ( -37.80, z-score = -5.94, R) >dp4.chr4_group3 6824024 101 + 11692001 CUUGUCGCUGCU-----GCUCGUUGUCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUUGCAACGGCAGCAG---------- ..(((.((((.(-----((..(((((((.......(((......)))..(((((..((((((......))).)))...))))).)))))))..))).)))).))).---------- ( -26.10, z-score = -2.69, R) >droPer1.super_1 8271929 104 + 10282868 CUUGUCGCUGCU-----GCUCGUUGUCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUUGCAACGGCAGCAGCAG------- ......((((((-----((.(((((.((......(((.((.(((((((....((((.((.....)).))))..))))))).)).))).....)))))))))))))))..------- ( -30.00, z-score = -3.28, R) >droWil1.scaffold_181038 347354 100 + 637489 ---CUUGCUCUC-----GUU-GUUGUCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGCGAAUGCAACUGACAACUAGCAACAACAACAGCCA------- ---...(((...-----(((-(((((..............(((.(((..(((.......)))..)))..)))(((....)))...........))))))))...)))..------- ( -15.30, z-score = -0.62, R) >droAna3.scaffold_12916 12088736 103 - 16180835 CUUGUCGCUGCU-----GCU-GUUGUCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUGGCAGCGGAGGAGCCAC------- ....((.(((((-----(((-(((((((.......(((......)))..(((((..((((((......))).)))...))))).))))))).)))))))).))......------- ( -27.60, z-score = -2.12, R) >droVir3.scaffold_12963 2146309 108 + 20206255 CAUCUCGCUGCUUAUUCGCC-GUUGCCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCUACUGACAACUGGCAGCGAAGACUACAG------- ..((((((((((......(.-((.((.(((((.((.((((..((((((((....))))).)))..)))).)).).)))))).)).)......))))))).)))......------- ( -21.10, z-score = -1.49, R) >droMoj3.scaffold_6500 27319327 108 + 32352404 CAUCUCGUUGCUUAUUCGUC-GUUGCCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUGGCAGAGAAGACAGCUA------- ..((((..((((..((.(((-(((((.(((((.((.((((..((((((((....))))).)))..)))).)).).))))))))).)))))..)))))))).........------- ( -20.80, z-score = -0.81, R) >droGri2.scaffold_15126 1201069 116 + 8399593 CUGCUCGCUGCUUAUUCGCCGUUUGCCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUGCCAACGGCAACAGAUCCCAGCAA ......((((.......((((((.((.........(((......)))..(((((..((((((......))).)))...))))).........)).)))))).........)))).. ( -20.09, z-score = -0.23, R) >consensus CUUGUCGCUGCU_____GCC_GUUGUCAUUCCUUUUAAUUUCAUUUAUUUUGCAAAAUACAAUUUAAUUUGAUGUGAAUGCAACUGACAACUGGCAGCGGCGGCAGCAG_______ ......((((.......(((.((((.((.....((((...(((.(((..(((.......)))..)))..)))..))))......)).)))).)))........))))......... (-11.13 = -11.62 + 0.49)

| Location | 11,378,908 – 11,379,011 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Shannon entropy | 0.28082 |
| G+C content | 0.37400 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -15.59 |
| Energy contribution | -15.42 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.748604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
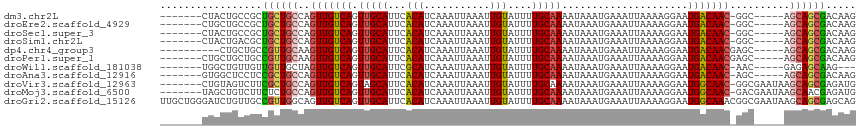
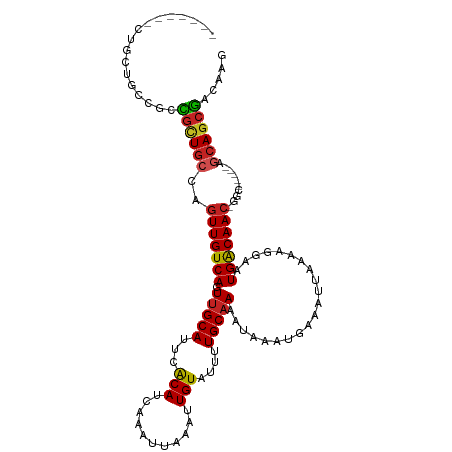
>dm3.chr2L 11378908 103 - 23011544 -------CUACUGCCGCUGCUGCCAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGACAAC-GGC-----AGCAGCGACAAG -------....((.((((((((((.(((((((.(((((...(((...........)))....))))).....................)))))))-)))-----))))))).)).. ( -35.25, z-score = -4.91, R) >droEre2.scaffold_4929 12576129 103 + 26641161 -------CUGCUGCCGCUGCUGCCAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGACAAC-GGC-----AGCAGCGACAAG -------....((.((((((((((.(((((((.(((((...(((...........)))....))))).....................)))))))-)))-----))))))).)).. ( -35.25, z-score = -4.25, R) >droSec1.super_3 6773339 103 - 7220098 -------CUACUGCCGCUGCUGCCAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGACAAC-GGC-----AGCAGCGACAAG -------....((.((((((((((.(((((((.(((((...(((...........)))....))))).....................)))))))-)))-----))))))).)).. ( -35.25, z-score = -4.91, R) >droSim1.chr2L 11185031 103 - 22036055 -------CUACUGACGCUGCUGCCAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGACAAC-GGC-----AGCAGCGACAAG -------....((.((((((((((.(((((((.(((((...(((...........)))....))))).....................)))))))-)))-----))))))).)).. ( -34.85, z-score = -5.10, R) >dp4.chr4_group3 6824024 101 - 11692001 ----------CUGCUGCCGUUGCAAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGACAACGAGC-----AGCAGCGACAAG ----------.((((((...(((..(((((((.(((((...(((...........)))....))))).....................)))))))..))-----)))))))..... ( -24.55, z-score = -1.53, R) >droPer1.super_1 8271929 104 - 10282868 -------CUGCUGCUGCCGUUGCAAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGACAACGAGC-----AGCAGCGACAAG -------.((((((((((((((((..((.(((.(((((...(((...........)))....)))))......)))........))..)).))))).))-----)))))))..... ( -28.10, z-score = -2.05, R) >droWil1.scaffold_181038 347354 100 - 637489 -------UGGCUGUUGUUGUUGCUAGUUGUCAGUUGCAUUCGCAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGACAAC-AAC-----GAGAGCAAG--- -------..(((.(((((((((((((((.(((.(((((...(((...........)))....)))))......))))))))........).))))-)))-----)).)))...--- ( -21.20, z-score = -0.65, R) >droAna3.scaffold_12916 12088736 103 + 16180835 -------GUGGCUCCUCCGCUGCCAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGACAAC-AGC-----AGCAGCGACAAG -------((.(((.....(((((..(((((((.(((((...(((...........)))....))))).....................)))))))-.))-----)))))).))... ( -25.05, z-score = -1.78, R) >droVir3.scaffold_12963 2146309 108 - 20206255 -------CUGUAGUCUUCGCUGCCAGUUGUCAGUAGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGGCAAC-GGCGAAUAAGCAGCGAGAUG -------.....((((.((((((...(((((.((.((((((......((((..(((.(((((....))))))))..)))).....)))).)).))-)))))....)))))))))). ( -25.10, z-score = -1.39, R) >droMoj3.scaffold_6500 27319327 108 - 32352404 -------UAGCUGUCUUCUCUGCCAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGGCAAC-GACGAAUAAGCAACGAGAUG -------.........(((((((...(((((.(((((((((......((((..(((.(((((....))))))))..)))).....)))).)))))-)))))....)))..)))).. ( -20.80, z-score = -0.58, R) >droGri2.scaffold_15126 1201069 116 - 8399593 UUGCUGGGAUCUGUUGCCGUUGGCAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGGCAAACGGCGAAUAAGCAGCGAGCAG ((((((.......(((((((((((....)))).((((((((......((((..(((.(((((....))))))))..)))).....)))).))))))))))).....)))))).... ( -25.50, z-score = 0.28, R) >consensus _______CUGCUGCCGCCGCUGCCAGUUGUCAGUUGCAUUCACAUCAAAUUAAAUUGUAUUUUGCAAAAUAAAUGAAAUUAAAAGGAAUGACAAC_GGC_____AGCAGCGACAAG .................((((((..(((((((.(((((...(((...........)))....))))).....................)))))))..........))))))..... (-15.59 = -15.42 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:17 2011