| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,193,140 – 14,193,235 |
| Length | 95 |
| Max. P | 0.981332 |

| Location | 14,193,140 – 14,193,235 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 67.16 |
| Shannon entropy | 0.60482 |
| G+C content | 0.41801 |
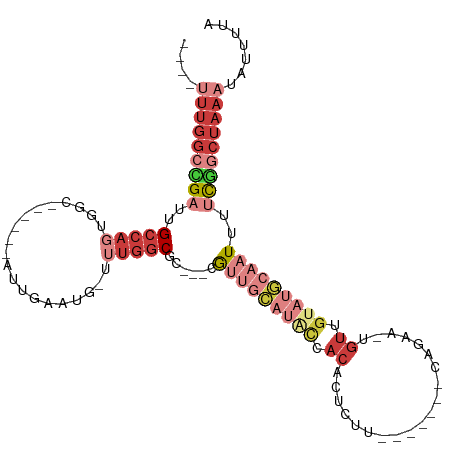
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -10.87 |
| Energy contribution | -14.14 |
| Covariance contribution | 3.27 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
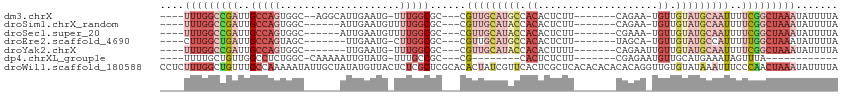
>dm3.chrX 14193140 95 - 22422827 ----UUUGGCCGAUUGCCAGUGGC--AGGCAUUGAAUG-UUUGGCGC---CGUUGCAUGCCACACUCUU-------CAGAA-UGUUGUAUGCAAUUUUCGGCUAAAUAUUUUA ----(((((((((......(((.(--((((((...)))-)))).)))---.(((((((((.(((.((..-------..)).-))).)))))))))..)))))))))....... ( -36.30, z-score = -3.51, R) >droSim1.chrX_random 3797961 92 - 5698898 ----UUUGGCCGAUUGCCAGUGGC------AUUGAAUGUUUUGGCGC---CGUUGCAUACCACACUCUU-------CAGAA-UGUUGUAUGCAAUUUUCGGCUAAAUAUUUUA ----(((((((((..(((((.(((------((...))))))))))..---.(((((((((.(((.((..-------..)).-))).)))))))))..)))))))))....... ( -34.40, z-score = -4.43, R) >droSec1.super_20 795086 92 - 1148123 ----UUUGGCCGAUUGCCAGUGGC------AUUGAAUGUUUUGGCGC---CGUUGCAUACCACACUCUU-------CGAAA-UGUUGUAUGCAAUUUUCGGCUAAAUAUUUUA ----(((((((((..(((((.(((------((...))))))))))..---.(((((((((.(((.....-------.....-))).)))))))))..)))))))))....... ( -31.20, z-score = -3.39, R) >droEre2.scaffold_4690 6010400 90 + 18748788 ----CUUGGCUGAUUGCCAGUAGC-------UUGAAUG-CUUGGCGC---CGUUGCAUGCCACACUCUU-------UAGCA-UGUUGUAUGCCAUUUUUGGCUAAAUAUUUUA ----.(((((.....)))))((((-------(.(((..-..(((((.---....((((((.........-------..)))-)))....))))).))).)))))......... ( -24.80, z-score = -0.66, R) >droYak2.chrX 8444052 91 - 21770863 ----UUUGGCCGAUUGCCAGUGGC-------UUGAAUG-UUUGGCGC---CGUUGCAUACCACACUUUU-------CAGAAUUGUUGUAUGCAAUUUUCGGCUAAAUAUUUUA ----(((((((((..(((((..((-------......)-))))))..---.(((((((((.(((.(((.-------..))).))).)))))))))..)))))))))....... ( -30.50, z-score = -3.38, R) >dp4.chrXL_group1e 7457696 77 + 12523060 ----UUUUGCUGUUGGCCUCUGGC-CAAAAAUUGUAUG-UUUGCCGC---CG--------CACUCUCUU-------CGAGAAUGUUGCAUGAAAUAGUUUA------------ ----....((((((((((...)))-)............-......((---.(--------((.((((..-------.)))).))).))....))))))...------------ ( -18.00, z-score = -1.14, R) >droWil1.scaffold_180588 10095 113 - 1294757 CCUCUUUGGCUGUUUGCCAAAAAUAUUGCUAUAUGUUACUCUCGCUCGCACACUAUCGUUCACUCGCUCACACACACACAGGUUGUGUAUAAAUUUCCCAACUAAAUAUUUUA ......((((.....))))((((((((................((......((....))......))..(((((.((....)))))))................)))))))). ( -12.90, z-score = 0.39, R) >consensus ____UUUGGCCGAUUGCCAGUGGC______AUUGAAUG_UUUGGCGC___CGUUGCAUACCACACUCUU_______CAGAA_UGUUGUAUGCAAUUUUCGGCUAAAUAUUUUA .....((((((((..(((((....................)))))......(((((((((.((....................)).)))))))))..))))))))........ (-10.87 = -14.14 + 3.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:35 2011