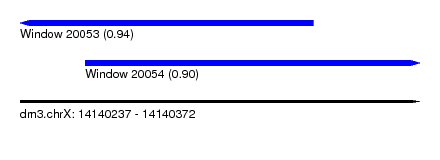
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,140,237 – 14,140,372 |
| Length | 135 |
| Max. P | 0.944879 |

| Location | 14,140,237 – 14,140,336 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 66.50 |
| Shannon entropy | 0.55306 |
| G+C content | 0.67649 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -24.74 |
| Energy contribution | -28.05 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14140237 99 - 22422827 GCCGGGAUUGAGCUUGGCCGCAGCACUGGCCUGGCCACCUGGACUGGCGACCGUACUGCGACCUGAUCUCGUACCCGGACCAGAUGCGCCAGCUGAAUC .(((((..((.(((.(((((......))))).)))))))))).((((((..((.....))..(((.((.((....)))).)))...))))))....... ( -35.80, z-score = -0.62, R) >droAna3.scaffold_13248 2998160 81 + 4840945 GCCGGGAUCG-GUGGGACUGACUGUAUGGGUUGGUCGGAUGGGCUUG-----GAGGUGUGUCCGGAUACUAUGCCGGGACUACUGUC------------ .((((.((.(-(((.(((..(((.....)))..)))(((((.(((..-----..))).)))))...)))))).))))(((....)))------------ ( -29.70, z-score = -1.45, R) >droSec1.super_20 743695 99 - 1148123 GCCGGGACGGGGCUUGGCUGCAGCGGUCGGCUGGCCACCUGGACUGGCGACCGUACUGCGACCUGGUCUCGUACCCGGACCCGAACCGCCAGCGGAGUC .(((((((((((((.((.(((((((((((.(..(((....)).)..)))))))..))))).)).)))))))).))))).......(((....))).... ( -53.50, z-score = -2.32, R) >droSim1.chrX 10859334 99 - 17042790 GCCGGGAUGGGGCUUGGCUGCAGCGGUCGGCUGGCCACCUGGAGUGGCGACCGUACUGCGACCUGGUCUCGUACCCGGACCCGAACCGCCAGCGGAGUC .(((((((((((((.((.(((((((((((.(...((....))...).))))))..))))).)).)))))))).))))).......(((....))).... ( -50.20, z-score = -1.35, R) >consensus GCCGGGAUGGGGCUUGGCUGCAGCGGUCGGCUGGCCACCUGGACUGGCGACCGUACUGCGACCUGAUCUCGUACCCGGACCAGAACCGCCAGCGGAGUC .(((((((((((((.((.(((((((((((.((((((....)).))))))))))..))))).)).)))))))).)))))..................... (-24.74 = -28.05 + 3.31)


| Location | 14,140,259 – 14,140,372 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.04 |
| Shannon entropy | 0.64808 |
| G+C content | 0.66164 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14140259 113 + 22422827 CGGGUACGAGAUCAGGUCGCAGUACGGUCGCCAGUCCAGGUGGCCAGGCCAGUGCUGCGGCCAAGCUCAAUCCCGGCGAGAAACGACAGCGACUACAGCGCCUGGACACGUUG ((((...(((....(((((((((((((((((((.......))))..)))).)))))))))))...)))...))))......((((.(((((.(....))).)))....)))). ( -46.80, z-score = -1.44, R) >droAna3.scaffold_13248 2998170 101 - 4840945 -----CCGGCAUAGUAUCCGGACACACCUCCAAGCCCAUCCGACCAACCCAUACAGUCAGUCCCAC-CGAUCCCGGCGAGAAGCGAUCCCAGCGACUAGUCAUGGAG------ -----((((........))))...........................((((((((((..((.(.(-((....))).).)).((.......)))))).)).))))..------ ( -21.20, z-score = -0.45, R) >droYak2.chrX 8393632 86 + 21770863 CGGGUUCGAAACCAGGUCGCAGUACGGCUG---------GUGGC------------------AAGCCCCAUCCCGGCGAGAAACGCCAGCGGCUACAGCGCCUGGACACAUUG ...........(((((.(((.((((.((((---------(((.(------------------..(((.......)))..)...))))))).).))).))))))))........ ( -35.70, z-score = -1.57, R) >droSec1.super_20 743717 113 + 1148123 CGGGUACGAGACCAGGUCGCAGUACGGUCGCCAGUCCAGGUGGCCAGCCGACCGCUGCAGCCAAGCCCCGUCCCGGCGAGAAACGCCAGCGGCUGCAGCGCCUGGACACGUUG (((((.........(((((..((..(((((((......))))))).)))))))(((((((((..(........)((((.....))))...))))))))))))))......... ( -50.90, z-score = -1.28, R) >droSim1.chrX 10859356 113 + 17042790 CGGGUACGAGACCAGGUCGCAGUACGGUCGCCACUCCAGGUGGCCAGCCGACCGCUGCAGCCAAGCCCCAUCCCGGCGAGAAACGCCAGCGGCUGCAGCGCCUGGACACGUUG (((((.........(((((..((..(((((((......))))))).)))))))(((((((((..(........)((((.....))))...))))))))))))))......... ( -50.90, z-score = -1.63, R) >consensus CGGGUACGAGACCAGGUCGCAGUACGGUCGCCAGUCCAGGUGGCCAGCCCACCGCUGCAGCCAAGCCCCAUCCCGGCGAGAAACGCCAGCGGCUACAGCGCCUGGACACGUUG ...........(((((.(((.....(((((((......))))))).............................((((.....))))..........))))))))........ (-19.13 = -19.98 + 0.85)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:32 2011