| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,129,819 – 14,129,916 |
| Length | 97 |
| Max. P | 0.825285 |

| Location | 14,129,819 – 14,129,916 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 60.19 |
| Shannon entropy | 0.85193 |
| G+C content | 0.44397 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -10.67 |
| Energy contribution | -10.59 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.25 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.825285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
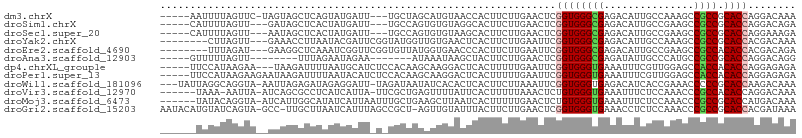
>dm3.chrX 14129819 97 + 22422827 -----AAUUUUAGUUC-UAGUAGCUCAGUAUGAUU---UGCUAGCAUGUAACCACUUCUUGAACUCGGUGGGCGAGACAUUGCCAAAGCCGCCGCACCAGGACAAA -----.......((((-(.((.(((.((((.....---)))))))....................((((.(((((....)))))...))))....)).)))))... ( -24.40, z-score = -0.41, R) >droSim1.chrX 10849225 95 + 17042790 -----CAUUUUAGUU---GAUAGCUCACUAUGAUU---UGCCAGUGUGUAGGCACUUCUUGAACUCGGUGGGCGAGACAUUGCCGAAGCCGCCGCACCAGGACAGA -----.....((((.---((....))))))...((---((((.(((((..(((.(.....)...((((..(........)..)))).)))..)))))..)).)))) ( -23.60, z-score = 0.73, R) >droSec1.super_20 733436 95 + 1148123 -----CAUUUUAGUU---AAUAGCUCACUAUGAUU---UGCCAGUGUGUAAGCACUUCUUGAACUCGGUGGGCGAGACAUUGCCGAAGCCGCCGCACCAGGAAAGA -----..........---....((((((..((...---...))..)))..)))..((((((....((((((((((....)))).....))))))...))))))... ( -21.10, z-score = 0.86, R) >droYak2.chrX 8383365 95 + 21770863 --------CUUAGUU---GAAACCUUAAUACGAUUCGGUAUGGUUGUGAACUCACUUCUUGAAUUCGGUGGGCGAGACAUUGCCAAAGCCGCCGCACCACGACAAA --------....(((---(..(((............))).((((.(((...(((.....)))...((((.(((((....)))))...)))).)))))))))))... ( -23.30, z-score = -0.53, R) >droEre2.scaffold_4690 5947845 95 - 18748788 --------UUUAGAU---GAAGGCUCAAAUCGGUUCGGUGUUAUGGUGAACCCACUUCUUGAAUUCGGUGGGCGAGACAUUGCCGAAGCCGCCACACCACGACAGA --------.......---...(((((((...((((((.(.....).))))))......))))....(((.(((((....)))))...))))))............. ( -22.80, z-score = 1.00, R) >droAna3.scaffold_12903 345611 86 - 802071 -----GUUUUUAGUU--------UUUAGAAUAGAA-------AUAAAUAAGCUACUUCUUGAACUCGGUGGGCGAGAUAUUGCCCAUGCCGCCGCACCAGGACAGG -----((((...(((--------((((...)))))-------))....))))..(((((((....((((((((((....))))))..))))......))))).)). ( -19.80, z-score = -0.57, R) >dp4.chrXL_group1e 8479825 98 + 12523060 -----UUCCAUAAGAA---UAAGAUUUUAAUGCAUCUCCACAAGCAAGGACUCACUUUUUGAAUUCGGUGGGUGAAAUUUCGUUGGAGCCACCACACCAGGAGAGA -----...........---...............(((((.((((.(((......))))))).....((((((((....(((....))).)))).)))).))))).. ( -22.50, z-score = -0.63, R) >droPer1.super_13 2199876 101 - 2293547 -----UUCCAUAAGAAGAAUAAGAUUUUAAUACAUCUCCACAAGCAAGGACUCACUUUUUGAAUUCGGUGGGUGAAAUUUCGUUGGAGCCACCACACCAGGAGAGA -----.............................(((((.((((.(((......))))))).....((((((((....(((....))).)))).)))).))))).. ( -22.50, z-score = -0.86, R) >droWil1.scaffold_181096 10598397 101 + 12416693 ---UAUUAGGCAGGUA-AAUUAGAGAUAGAGGAUU-UAGAUAAUAUCACACUCACUUCUUAAAUUCGGUGGGUGAGACAUCACCGAAACCCCCGCACCAAGACAAA ---.....((..(((.-.....((((..(((....-..............)))...))))...((((((((........))))))))))).))............. ( -18.87, z-score = -0.50, R) >droVir3.scaffold_12970 10831094 97 - 11907090 ------UAAA-AAUUA-AUCAGCGCCUCAUCAUUA-UUCGCUGAGUUUUAUUCACUUUUUAAACUCUGUGGGUGAAAUUUCUCCAAACCCGCCACACCAGGACAAA ------....-.....-.((((((...........-..))))))(((((((((((............)))))))))))............(((......)).)... ( -16.12, z-score = -1.17, R) >droMoj3.scaffold_6473 874627 99 - 16943266 ------UAUACAGGUA-AUCAUUGGCAUAUCAUUAAUUUGCUGAAGCUUAAUCACUUUUUGAACUCUGUGGGUGAAAUUUCUCCAAACCCGCCGCACCAUGACAAA ------....(((((.-....(..(((...........)))..).((....(((.....))).....((((((.............)))))).))))).))..... ( -17.52, z-score = -0.11, R) >droGri2.scaffold_15203 11957900 103 + 11997470 AAUACAUGUAUCAGUA-GCC-UUGCUUAAUCAUUUAGCCGCU-AGUUGUAUUUACUUCUUGAACUCGGUGGGUGAAACCUCUCCAAACCCGCCGCACCACGAUAAA ........((((..((-((.-..(((.........))).)))-).....................((((((((.............))))))))......)))).. ( -20.72, z-score = -0.77, R) >consensus ______UUUUUAGUUA__AAAAGCUCAAAAUGAUU_U_UGCUAGAGUGUACUCACUUCUUGAACUCGGUGGGCGAGACAUCGCCGAAGCCGCCGCACCAGGACAAA ..................................................................((((((((...............)))).))))........ (-10.67 = -10.59 + -0.08)

| Location | 14,129,819 – 14,129,916 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 60.19 |
| Shannon entropy | 0.85193 |
| G+C content | 0.44397 |
| Mean single sequence MFE | -23.09 |
| Consensus MFE | -9.63 |
| Energy contribution | -9.96 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.52 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
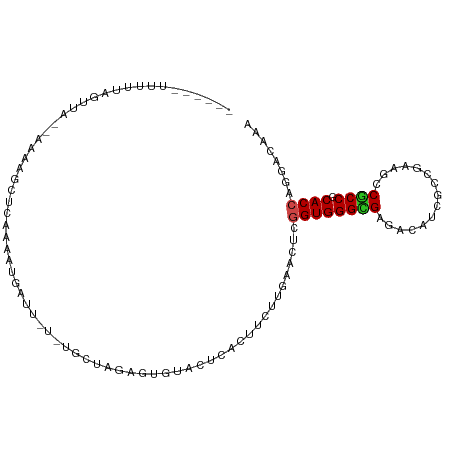
>dm3.chrX 14129819 97 - 22422827 UUUGUCCUGGUGCGGCGGCUUUGGCAAUGUCUCGCCCACCGAGUUCAAGAAGUGGUUACAUGCUAGCA---AAUCAUACUGAGCUACUA-GAACUAAAAUU----- ((((..((((((.(((((....(((...)))))))))))).))..))))...(((((...((.((((.---..((.....)))))).))-.))))).....----- ( -22.70, z-score = 0.43, R) >droSim1.chrX 10849225 95 - 17042790 UCUGUCCUGGUGCGGCGGCUUCGGCAAUGUCUCGCCCACCGAGUUCAAGAAGUGCCUACACACUGGCA---AAUCAUAGUGAGCUAUC---AACUAAAAUG----- .......(((((.((((((....)).......)))))))))((((((.((..((((........))))---..))....))))))...---..........----- ( -29.51, z-score = -1.91, R) >droSec1.super_20 733436 95 - 1148123 UCUUUCCUGGUGCGGCGGCUUCGGCAAUGUCUCGCCCACCGAGUUCAAGAAGUGCUUACACACUGGCA---AAUCAUAGUGAGCUAUU---AACUAAAAUG----- .......(((((.((((((....)).......)))))))))((((((.((..((((........))))---..))....))))))...---..........----- ( -27.11, z-score = -1.57, R) >droYak2.chrX 8383365 95 - 21770863 UUUGUCGUGGUGCGGCGGCUUUGGCAAUGUCUCGCCCACCGAAUUCAAGAAGUGAGUUCACAACCAUACCGAAUCGUAUUAAGGUUUC---AACUAAG-------- ......((((((.(((((....(((...))))))))))))(((((((.....)))))))))((((((((......))))...))))..---.......-------- ( -22.90, z-score = -0.36, R) >droEre2.scaffold_4690 5947845 95 + 18748788 UCUGUCGUGGUGUGGCGGCUUCGGCAAUGUCUCGCCCACCGAAUUCAAGAAGUGGGUUCACCAUAACACCGAACCGAUUUGAGCCUUC---AUCUAAA-------- ..((...(((((.((((((....)).......)))))))))....))(((.(.(((((((...................))))))).)---.)))...-------- ( -25.32, z-score = -0.12, R) >droAna3.scaffold_12903 345611 86 + 802071 CCUGUCCUGGUGCGGCGGCAUGGGCAAUAUCUCGCCCACCGAGUUCAAGAAGUAGCUUAUUUAU-------UUCUAUUCUAAA--------AACUAAAAAC----- ..((..((((((.(((((.(((.....))).))))))))).))..))((((((((.....))))-------))))........--------..........----- ( -21.40, z-score = -1.55, R) >dp4.chrXL_group1e 8479825 98 - 12523060 UCUCUCCUGGUGUGGUGGCUCCAACGAAAUUUCACCCACCGAAUUCAAAAAGUGAGUCCUUGCUUGUGGAGAUGCAUUAAAAUCUUA---UUCUUAUGGAA----- ..((((((((((.(((((.............))))))))))........(((..(....)..)))..)))))...............---...........----- ( -21.42, z-score = -0.40, R) >droPer1.super_13 2199876 101 + 2293547 UCUCUCCUGGUGUGGUGGCUCCAACGAAAUUUCACCCACCGAAUUCAAAAAGUGAGUCCUUGCUUGUGGAGAUGUAUUAAAAUCUUAUUCUUCUUAUGGAA----- ..((((((((((.(((((.............))))))))))........(((..(....)..)))..)))))...............((((......))))----- ( -22.02, z-score = -0.75, R) >droWil1.scaffold_181096 10598397 101 - 12416693 UUUGUCUUGGUGCGGGGGUUUCGGUGAUGUCUCACCCACCGAAUUUAAGAAGUGAGUGUGAUAUUAUCUA-AAUCCUCUAUCUCUAAUU-UACCUGCCUAAUA--- ........((((.((((((((.(((((((((.(((.(((............))).))).))))))))).)-))))))))))).......-.............--- ( -27.90, z-score = -2.46, R) >droVir3.scaffold_12970 10831094 97 + 11907090 UUUGUCCUGGUGUGGCGGGUUUGGAGAAAUUUCACCCACAGAGUUUAAAAAGUGAAUAAAACUCAGCGAA-UAAUGAUGAGGCGCUGAU-UAAUU-UUUA------ ((((..((..(((((..((..(......)..))..))))).))..)))).............((((((..-(.......)..)))))).-.....-....------ ( -16.70, z-score = 1.02, R) >droMoj3.scaffold_6473 874627 99 + 16943266 UUUGUCAUGGUGCGGCGGGUUUGGAGAAAUUUCACCCACAGAGUUCAAAAAGUGAUUAAGCUUCAGCAAAUUAAUGAUAUGCCAAUGAU-UACCUGUAUA------ .........((((((.((((..(((....))).))))..............(((((((.((.(((.........)))...))...))))-))))))))).------ ( -17.60, z-score = 1.05, R) >droGri2.scaffold_15203 11957900 103 - 11997470 UUUAUCGUGGUGCGGCGGGUUUGGAGAGGUUUCACCCACCGAGUUCAAGAAGUAAAUACAACU-AGCGGCUAAAUGAUUAAGCAA-GGC-UACUGAUACAUGUAUU ..(((((((((((.(((..(((((.(.((.....))).)))))..)....(((.......)))-.)).))....((......)).-.))-))).))))........ ( -22.50, z-score = 0.38, R) >consensus UCUGUCCUGGUGCGGCGGCUUCGGCAAUGUCUCACCCACCGAGUUCAAGAAGUGAGUACACACUAGCA_A_AAUCAUUUUGAGCUUUU__UAACUAAAAA______ .......(((((.(((((.............))))))))))................................................................. ( -9.63 = -9.96 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:30 2011